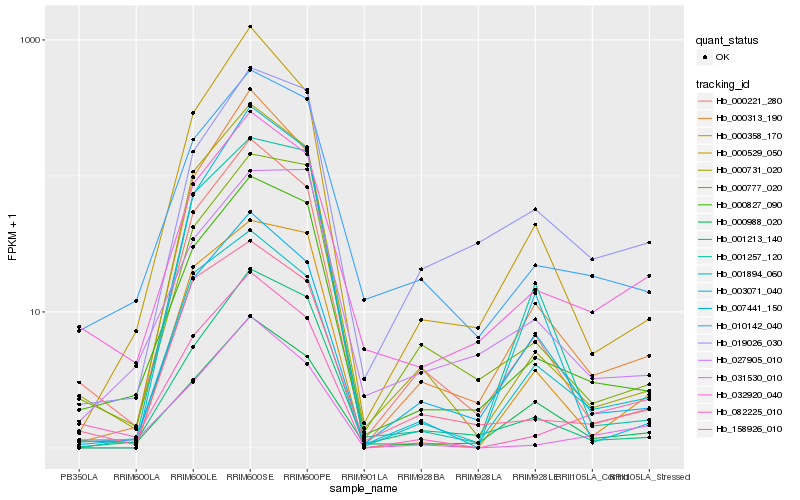
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000827_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 2 |
Hb_010142_040 |
0.0677178143 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 3 |
Hb_001213_140 |
0.0722825702 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 4 |
Hb_000777_020 |
0.0903319087 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 5 |
Hb_000221_280 |
0.1039659872 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 6 |
Hb_032920_040 |
0.1086123648 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 7 |
Hb_000731_020 |
0.1150160274 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 8 |
Hb_027905_010 |
0.1173509392 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 9 |
Hb_000358_170 |
0.1309472046 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 10 |
Hb_001257_120 |
0.1319784108 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 11 |
Hb_000313_190 |
0.1353679721 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 12 |
Hb_019026_030 |
0.1360525933 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 13 |
Hb_158926_010 |
0.1360731014 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 14 |
Hb_007441_150 |
0.13732976 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 15 |
Hb_000529_050 |
0.1384789162 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 16 |
Hb_003071_040 |
0.1404136425 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 17 |
Hb_001894_060 |
0.1418653173 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 18 |
Hb_000988_020 |
0.1429001343 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 19 |
Hb_082225_010 |
0.1450276185 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 20 |
Hb_031530_010 |
0.1484366621 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |