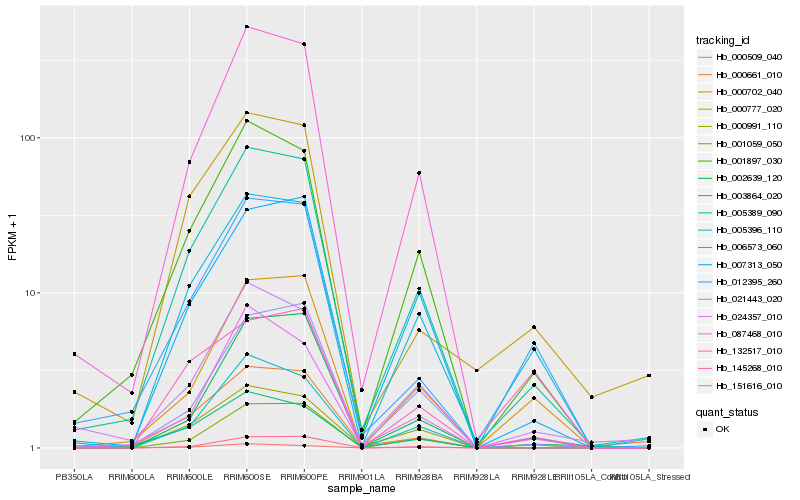
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005389_090 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 2 |
Hb_132517_010 |
0.0694639183 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 3 |
Hb_001897_030 |
0.072824792 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 4 |
Hb_006573_060 |
0.0950887385 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 5 |
Hb_000991_110 |
0.1045295169 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 6 |
Hb_007313_050 |
0.1138024893 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 7 |
Hb_005396_110 |
0.1173384849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001059_050 |
0.1182737825 |
- |
- |
PREDICTED: putative F-box/LRR-repeat protein At3g18150 [Jatropha curcas] |
| 9 |
Hb_024357_010 |
0.1225088063 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 10 |
Hb_012395_260 |
0.1269140811 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 11 |
Hb_021443_020 |
0.1271298624 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 12 |
Hb_002639_120 |
0.127626834 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_003864_020 |
0.1277718451 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_000777_020 |
0.1288255771 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 15 |
Hb_145268_010 |
0.1290779248 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 16 |
Hb_151616_010 |
0.1297912975 |
- |
- |
- |
| 17 |
Hb_087468_010 |
0.1310152009 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 18 |
Hb_000702_040 |
0.1332709052 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 19 |
Hb_000509_040 |
0.1341589339 |
- |
- |
hypothetical protein VITISV_009716 [Vitis vinifera] |
| 20 |
Hb_000661_010 |
0.1354508354 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |