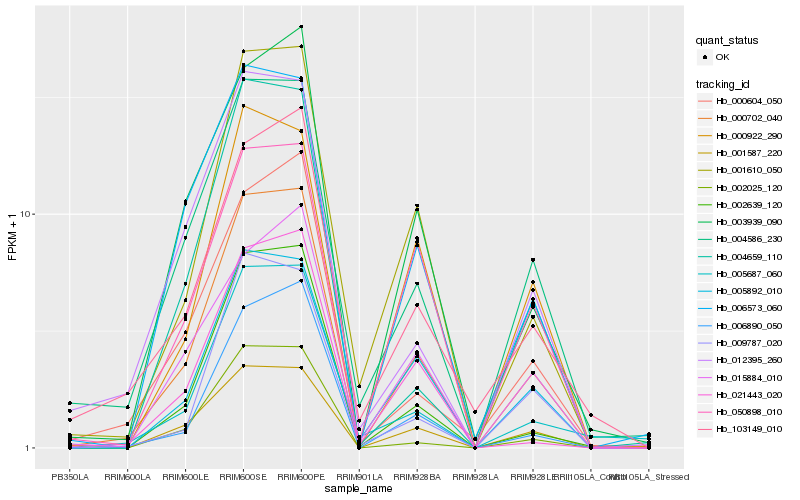
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000702_040 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_001610_050 |
0.097501864 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 3 |
Hb_004586_230 |
0.101619026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001587_220 |
0.1029159402 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_002639_120 |
0.1050969252 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_006890_050 |
0.1073871908 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Jatropha curcas] |
| 7 |
Hb_005892_010 |
0.1104725617 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 8 |
Hb_003939_090 |
0.11280494 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_006573_060 |
0.1131758735 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 10 |
Hb_012395_260 |
0.1192189696 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000604_050 |
0.1214097885 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 12 |
Hb_002025_120 |
0.121974021 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 13 |
Hb_009787_020 |
0.125847406 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 14 |
Hb_103149_010 |
0.1275104893 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 15 |
Hb_005687_060 |
0.1285208103 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000922_290 |
0.1286344745 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 17 |
Hb_015884_010 |
0.1289143929 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004659_110 |
0.1294544093 |
- |
- |
PREDICTED: BURP domain-containing protein 3-like [Jatropha curcas] |
| 19 |
Hb_021443_020 |
0.130274259 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 20 |
Hb_050898_010 |
0.1320690024 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |