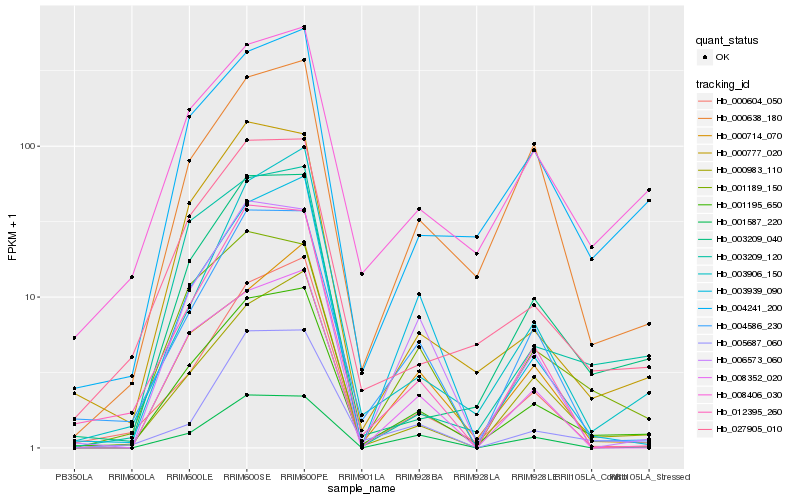
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_650 |
0.0 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 2 |
Hb_027905_010 |
0.1126742956 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 3 |
Hb_003209_040 |
0.1144070714 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 4 |
Hb_004586_230 |
0.1161746714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000983_110 |
0.1201700246 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 6 |
Hb_000777_020 |
0.1206389445 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 7 |
Hb_004241_200 |
0.1220152212 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 8 |
Hb_008406_030 |
0.122346617 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 9 |
Hb_000604_050 |
0.1240792395 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 10 |
Hb_012395_260 |
0.1274331749 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006573_060 |
0.1287940932 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 12 |
Hb_003939_090 |
0.1346193111 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_008352_020 |
0.1347082918 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 14 |
Hb_005687_060 |
0.1360389173 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001587_220 |
0.1366303284 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_001189_150 |
0.1376660871 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 17 |
Hb_000638_180 |
0.1405400546 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 18 |
Hb_003209_120 |
0.1440504905 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 19 |
Hb_000714_070 |
0.1452619788 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 20 |
Hb_003906_150 |
0.1470666061 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |