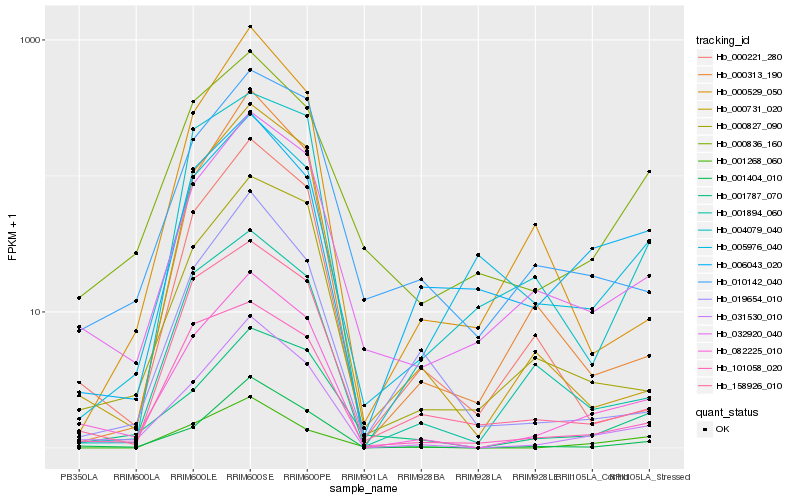
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_082225_010 |
0.0 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 2 |
Hb_031530_010 |
0.0518093966 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 3 |
Hb_001404_010 |
0.1077246362 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 4 |
Hb_032920_040 |
0.113882669 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 5 |
Hb_000827_090 |
0.1450276185 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 6 |
Hb_010142_040 |
0.1490208584 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 7 |
Hb_158926_010 |
0.1498340351 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 8 |
Hb_001787_070 |
0.1507943292 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Camelina sativa] |
| 9 |
Hb_101058_020 |
0.152798749 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 10 |
Hb_000836_160 |
0.1540331406 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 11 |
Hb_001894_060 |
0.1555620166 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 12 |
Hb_000221_280 |
0.1627254659 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 13 |
Hb_000731_020 |
0.1638661153 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 14 |
Hb_000313_190 |
0.1689854639 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 15 |
Hb_019654_010 |
0.1780901158 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 16 |
Hb_004079_040 |
0.1807425018 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 17 |
Hb_001268_060 |
0.1808051297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005976_040 |
0.1813390771 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 19 |
Hb_000529_050 |
0.1820467282 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 20 |
Hb_006043_020 |
0.1828915477 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |