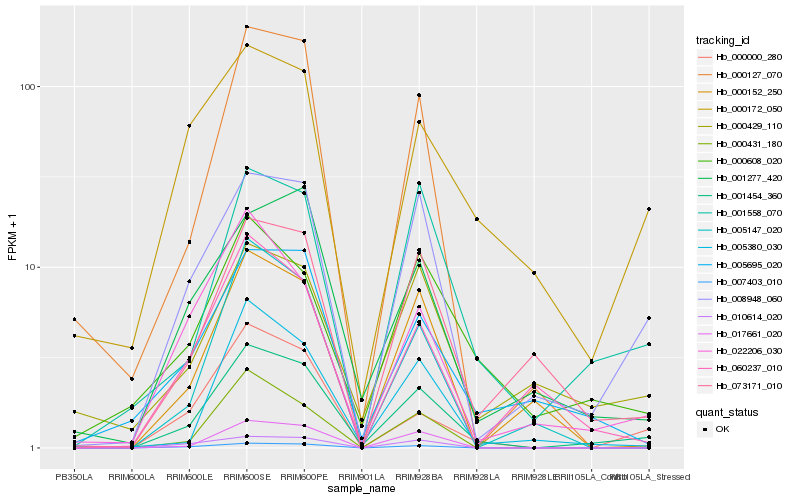
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_360 |
0.0 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 2 |
Hb_008948_060 |
0.1573116413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001558_070 |
0.1655728481 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 4 |
Hb_000000_280 |
0.171065665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007403_010 |
0.1778618505 |
- |
- |
hypothetical protein JCGZ_11090 [Jatropha curcas] |
| 6 |
Hb_073171_010 |
0.1805147355 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 7 |
Hb_017661_020 |
0.1809244811 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 8 |
Hb_005380_030 |
0.1848461508 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase B120 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005695_020 |
0.1878231371 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 10 |
Hb_022206_030 |
0.1890899369 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 11 |
Hb_000127_070 |
0.189430759 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 12 |
Hb_001277_420 |
0.1904597983 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 13 |
Hb_000431_180 |
0.191401178 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 14 |
Hb_000608_020 |
0.1944113134 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 15 |
Hb_005147_020 |
0.1988494571 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 16 |
Hb_000172_050 |
0.200499459 |
- |
- |
PREDICTED: uncharacterized protein LOC105650611 [Jatropha curcas] |
| 17 |
Hb_000429_110 |
0.2008826993 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 18 |
Hb_000152_250 |
0.202308891 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 19 |
Hb_010614_020 |
0.2043336235 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 20 |
Hb_060237_010 |
0.2049914876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |