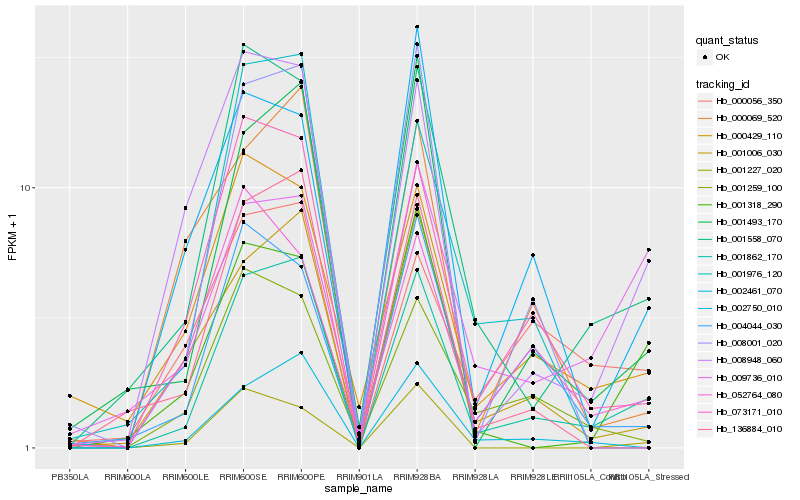
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001862_170 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_001558_070 |
0.1404904344 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 3 |
Hb_073171_010 |
0.1542039943 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 4 |
Hb_008948_060 |
0.1656212601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001006_030 |
0.1659795957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001493_170 |
0.1704273974 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 7 |
Hb_002461_070 |
0.1826382389 |
- |
- |
gibberellin 2-oxidase, putative [Ricinus communis] |
| 8 |
Hb_000429_110 |
0.187542239 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 9 |
Hb_001259_100 |
0.1882942087 |
- |
- |
chloride channel-like family protein [Populus trichocarpa] |
| 10 |
Hb_004044_030 |
0.1910596436 |
- |
- |
PREDICTED: uncharacterized protein LOC105639335 [Jatropha curcas] |
| 11 |
Hb_001318_290 |
0.193839342 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 12 |
Hb_136884_010 |
0.1940511193 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 13 |
Hb_008001_020 |
0.1983197129 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 14 |
Hb_000056_350 |
0.20108274 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 15 |
Hb_000069_520 |
0.2055180858 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 16 |
Hb_052764_080 |
0.2055342175 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001976_120 |
0.2060640411 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_002750_010 |
0.2070376199 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 19 |
Hb_009736_010 |
0.2090413437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001227_020 |
0.2099281818 |
- |
- |
- |