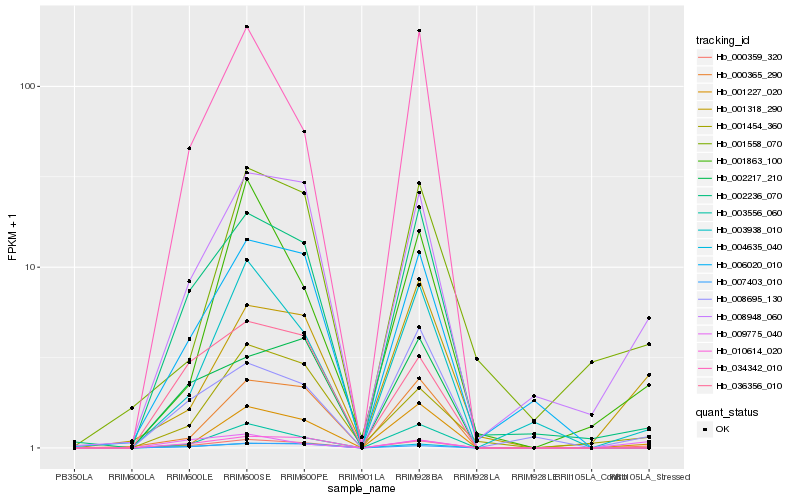
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_320 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 2 |
Hb_008948_060 |
0.1328662114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001318_290 |
0.1720629727 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 4 |
Hb_001227_020 |
0.1761130666 |
- |
- |
- |
| 5 |
Hb_002236_070 |
0.1808229146 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 6 |
Hb_003938_010 |
0.2040247834 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 7 |
Hb_008695_130 |
0.2099026844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004635_040 |
0.2119951922 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 9 |
Hb_001454_360 |
0.2141817102 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET17 [Jatropha curcas] |
| 10 |
Hb_010614_020 |
0.214730777 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 11 |
Hb_009775_040 |
0.2202424343 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Vitis vinifera] |
| 12 |
Hb_003556_060 |
0.2235300809 |
- |
- |
- |
| 13 |
Hb_001558_070 |
0.2300371314 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 14 |
Hb_006020_010 |
0.230588273 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 15 |
Hb_007403_010 |
0.2311129987 |
- |
- |
hypothetical protein JCGZ_11090 [Jatropha curcas] |
| 16 |
Hb_000365_290 |
0.2348510397 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_034342_010 |
0.2366924928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_036356_010 |
0.2367435452 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_002217_210 |
0.2373164201 |
- |
- |
- |
| 20 |
Hb_001863_100 |
0.2386531581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |