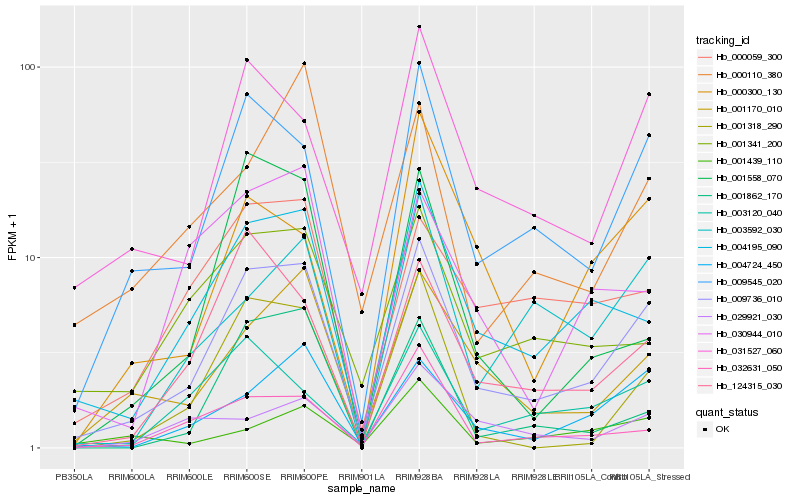
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009736_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009545_020 |
0.1367163106 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 3 |
Hb_004195_090 |
0.1524470984 |
- |
- |
PREDICTED: probable mitochondrial chaperone bcs1 [Jatropha curcas] |
| 4 |
Hb_031527_060 |
0.1613873096 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 5 |
Hb_001170_010 |
0.1693347453 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_004724_450 |
0.1754796332 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 7 |
Hb_124315_030 |
0.1924657729 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 8 |
Hb_001318_290 |
0.1947503947 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001341_200 |
0.195829872 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 10 |
Hb_000059_300 |
0.1959931723 |
- |
- |
caspase, putative [Ricinus communis] |
| 11 |
Hb_003120_040 |
0.2048249169 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 12 |
Hb_032631_050 |
0.2058050794 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_003592_030 |
0.2064206468 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 14 |
Hb_001862_170 |
0.2090413437 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_030944_010 |
0.2131749691 |
- |
- |
PREDICTED: uncharacterized protein LOC105633455 [Jatropha curcas] |
| 16 |
Hb_000110_380 |
0.2167622281 |
- |
- |
PREDICTED: uncharacterized protein LOC105642019 [Jatropha curcas] |
| 17 |
Hb_001558_070 |
0.2213338096 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 18 |
Hb_000300_130 |
0.2214282096 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 19 |
Hb_001439_110 |
0.2234565404 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |
| 20 |
Hb_029921_030 |
0.2242519726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |