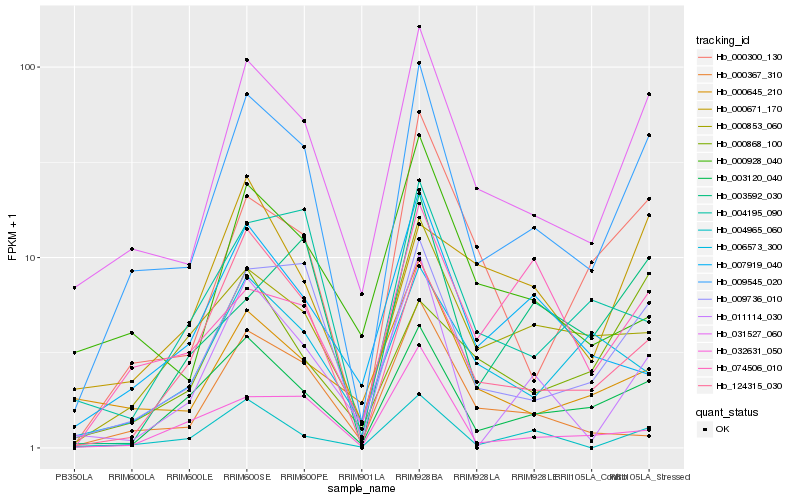
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009545_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 2 |
Hb_031527_060 |
0.1017063897 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 3 |
Hb_009736_010 |
0.1367163106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003120_040 |
0.1653114231 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 5 |
Hb_124315_030 |
0.1885373619 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 6 |
Hb_000853_060 |
0.1919144989 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004965_060 |
0.1958547023 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U4/U6 small nuclear ribonucleoprotein PRP4-like protein [Jatropha curcas] |
| 8 |
Hb_000300_130 |
0.198249223 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 9 |
Hb_011114_030 |
0.2045866591 |
- |
- |
PREDICTED: probable carboxylesterase 15 [Jatropha curcas] |
| 10 |
Hb_000645_210 |
0.2179027035 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_032631_050 |
0.223769058 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 12 |
Hb_004195_090 |
0.2239305789 |
- |
- |
PREDICTED: probable mitochondrial chaperone bcs1 [Jatropha curcas] |
| 13 |
Hb_000928_040 |
0.2241644285 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_003592_030 |
0.2284295905 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 15 |
Hb_006573_300 |
0.2295320498 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_007919_040 |
0.2309760731 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 17 |
Hb_000367_310 |
0.2325383036 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 18 |
Hb_074506_010 |
0.2351207839 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 19 |
Hb_000868_100 |
0.2360704562 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 20 |
Hb_000671_170 |
0.2362300459 |
- |
- |
kinase, putative [Ricinus communis] |