| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
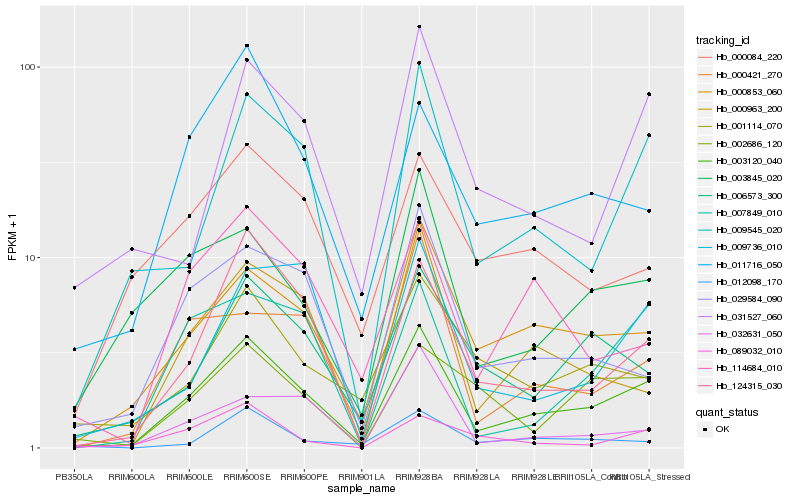
Hb_003120_040 |
0.0 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 2 |
Hb_000853_060 |
0.1517449207 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_124315_030 |
0.1622139439 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 4 |
Hb_009545_020 |
0.1653114231 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 5 |
Hb_000963_200 |
0.1855721989 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 6 |
Hb_007849_010 |
0.1873404374 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 7 |
Hb_006573_300 |
0.1893320903 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_029584_090 |
0.1933125489 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 9 |
Hb_011716_050 |
0.1960629793 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 10 |
Hb_003845_020 |
0.1960822285 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000421_270 |
0.1976373919 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 12 |
Hb_031527_060 |
0.2019031818 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 13 |
Hb_114684_010 |
0.2046092187 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 14 |
Hb_009736_010 |
0.2048249169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001114_070 |
0.2067002295 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g58400 [Jatropha curcas] |
| 16 |
Hb_002686_120 |
0.2116856498 |
- |
- |
short-chain type dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_032631_050 |
0.2144743995 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 18 |
Hb_000084_220 |
0.2170806379 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 19 |
Hb_089032_010 |
0.2176971661 |
- |
- |
PREDICTED: flotillin-like protein 3 [Jatropha curcas] |
| 20 |
Hb_012098_170 |
0.2178590888 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_16874 [Jatropha curcas] |