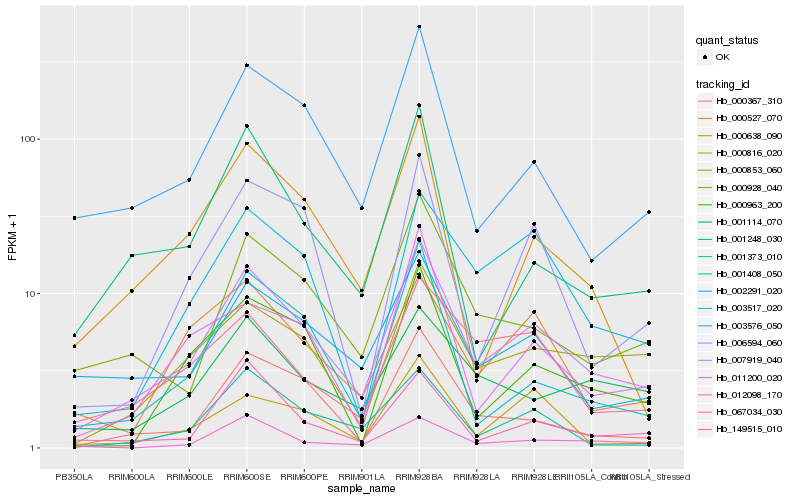
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007919_040 |
0.0 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 2 |
Hb_003576_050 |
0.1419653897 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 3 |
Hb_000853_060 |
0.1441967067 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003517_020 |
0.1458263909 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 5 |
Hb_000928_040 |
0.1491391803 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_000367_310 |
0.1510464402 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 7 |
Hb_002291_020 |
0.1549033074 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001373_010 |
0.1589296227 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_149515_010 |
0.1653647163 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 10 |
Hb_000638_090 |
0.1671843636 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_001114_070 |
0.1672026932 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g58400 [Jatropha curcas] |
| 12 |
Hb_001408_050 |
0.1675240764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_067034_030 |
0.1690381691 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_000527_070 |
0.1700595306 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 15 |
Hb_001248_030 |
0.1705178821 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_006594_060 |
0.1725178045 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 17 |
Hb_000963_200 |
0.1728887786 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 18 |
Hb_011200_020 |
0.1752716799 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 19 |
Hb_012098_170 |
0.1767300862 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_16874 [Jatropha curcas] |
| 20 |
Hb_000816_020 |
0.1769040865 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |