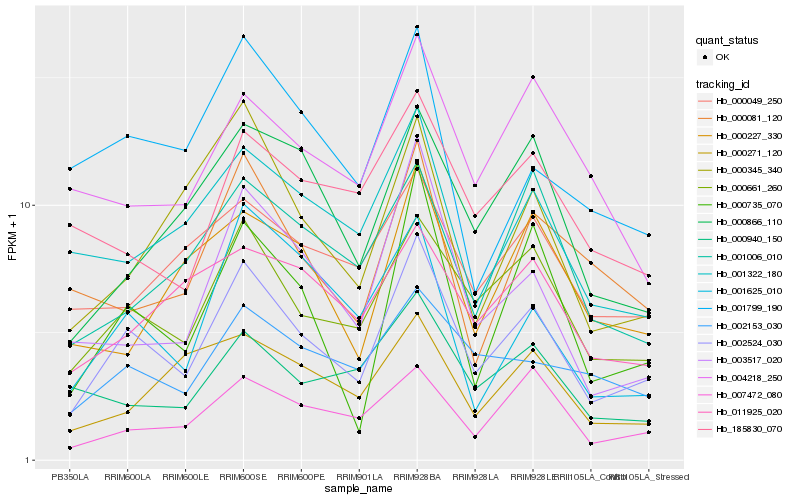
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002524_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000661_260 |
0.1046303889 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 3 |
Hb_000735_070 |
0.1433763696 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 4 |
Hb_007472_080 |
0.1434346758 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 5 |
Hb_002153_030 |
0.1474686485 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 6 |
Hb_003517_020 |
0.1481256764 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 7 |
Hb_001322_180 |
0.1492117961 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 8 |
Hb_000866_110 |
0.154223654 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001006_010 |
0.1570546082 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 10 |
Hb_000345_340 |
0.1583362951 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 11 |
Hb_000049_250 |
0.160701191 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000081_120 |
0.1607281626 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 13 |
Hb_001625_010 |
0.1614534978 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 14 |
Hb_011925_020 |
0.1633111515 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_000940_150 |
0.1640109218 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 16 |
Hb_001799_190 |
0.1647597964 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 17 |
Hb_185830_070 |
0.1668662554 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000271_120 |
0.1671008008 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 19 |
Hb_000227_330 |
0.1675167001 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 20 |
Hb_004218_250 |
0.1678665748 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |