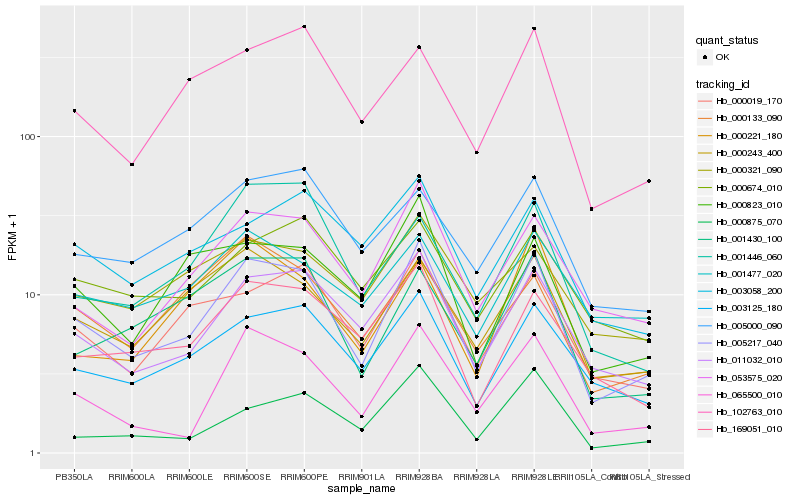
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005217_040 |
0.0 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011032_010 |
0.1031900979 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 3 |
Hb_003125_180 |
0.1152462913 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 4 |
Hb_053575_020 |
0.1179463607 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 5 |
Hb_065500_010 |
0.1191623013 |
- |
- |
- |
| 6 |
Hb_000019_170 |
0.1252085575 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 7 |
Hb_169051_010 |
0.1256820751 |
- |
- |
JHL06B08.1 [Jatropha curcas] |
| 8 |
Hb_000243_400 |
0.1264798205 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 9 |
Hb_102763_010 |
0.1397544148 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 10 |
Hb_000823_010 |
0.1402417737 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 11 |
Hb_001430_100 |
0.1445457797 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_005000_090 |
0.1445508622 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 13 |
Hb_003058_200 |
0.1448320641 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 14 |
Hb_000674_010 |
0.1479244862 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000133_090 |
0.1487096531 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 16 |
Hb_000321_090 |
0.1515217654 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 17 |
Hb_001477_020 |
0.1526149936 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 18 |
Hb_001446_060 |
0.153860448 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000221_180 |
0.1544443079 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 20 |
Hb_000875_070 |
0.155681983 |
- |
- |
- |