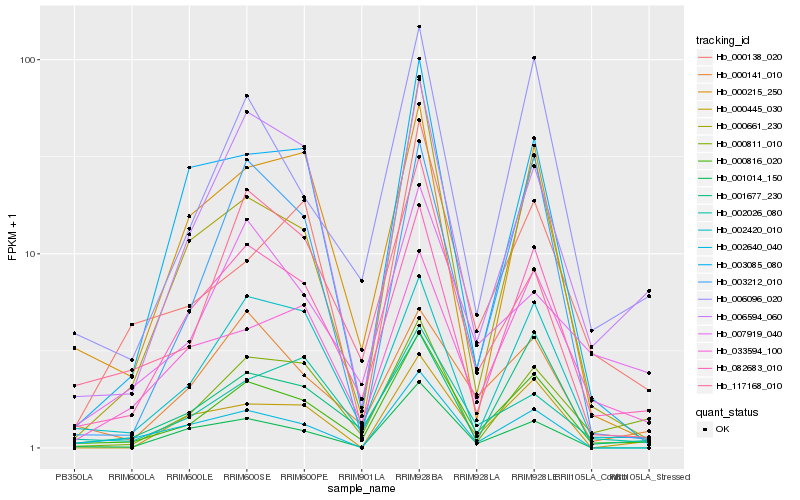
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_082683_010 |
0.1083010072 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_006096_020 |
0.1279725629 |
- |
- |
Os06g0343500 [Oryza sativa Japonica Group] |
| 4 |
Hb_001677_230 |
0.144470697 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002420_010 |
0.1511073158 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 6 |
Hb_033594_100 |
0.1550643469 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000811_010 |
0.1562877077 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 8 |
Hb_000661_230 |
0.1584521503 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_006594_060 |
0.1600201797 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 10 |
Hb_001014_150 |
0.1615349949 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000141_010 |
0.1662858283 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 12 |
Hb_000138_020 |
0.1696799994 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 13 |
Hb_002640_040 |
0.1718790325 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_002026_080 |
0.1726388143 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 15 |
Hb_000445_030 |
0.1736269466 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 16 |
Hb_000215_250 |
0.174099842 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_117168_010 |
0.1764003298 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 18 |
Hb_003212_010 |
0.1764516823 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 19 |
Hb_007919_040 |
0.1769040865 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 20 |
Hb_003085_080 |
0.1780408413 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |