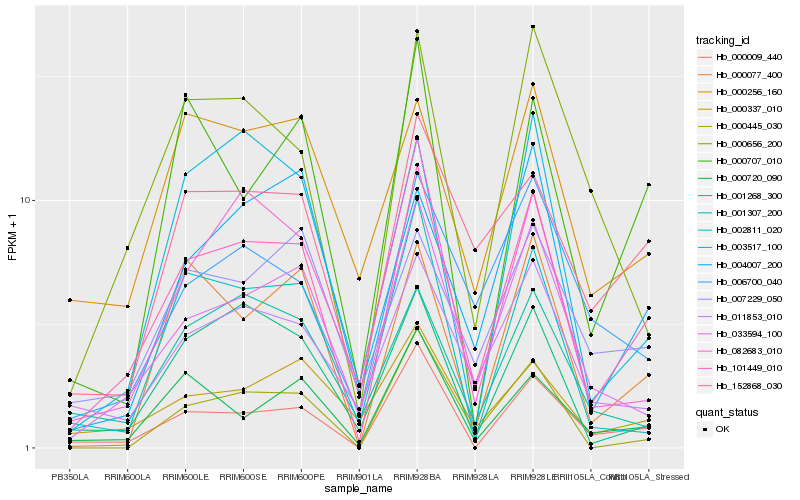
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101449_010 |
0.0 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 2 |
Hb_033594_100 |
0.14344567 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000009_440 |
0.1547213078 |
- |
- |
PREDICTED: uncharacterized protein LOC105639617 [Jatropha curcas] |
| 4 |
Hb_001268_300 |
0.1609307366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_082683_010 |
0.1652809302 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_011853_010 |
0.1655586837 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 7 |
Hb_152868_030 |
0.1660019017 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 8 |
Hb_000077_400 |
0.1703422645 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000720_090 |
0.1722807126 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 10 |
Hb_004007_200 |
0.1743682303 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 11 |
Hb_000337_010 |
0.1747771152 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000656_200 |
0.1768115888 |
- |
- |
PREDICTED: uncharacterized protein LOC105631373 [Jatropha curcas] |
| 13 |
Hb_003517_100 |
0.1775271017 |
transcription factor |
TF Family: GRAS |
- |
| 14 |
Hb_002811_020 |
0.1781009625 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 15 |
Hb_000707_010 |
0.1794274685 |
- |
- |
hypothetical protein JCGZ_25098 [Jatropha curcas] |
| 16 |
Hb_006700_040 |
0.1798199706 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 17 |
Hb_007229_050 |
0.1803395489 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000256_160 |
0.1810815885 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 19 |
Hb_000445_030 |
0.1818571915 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 20 |
Hb_001307_200 |
0.1828033796 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |