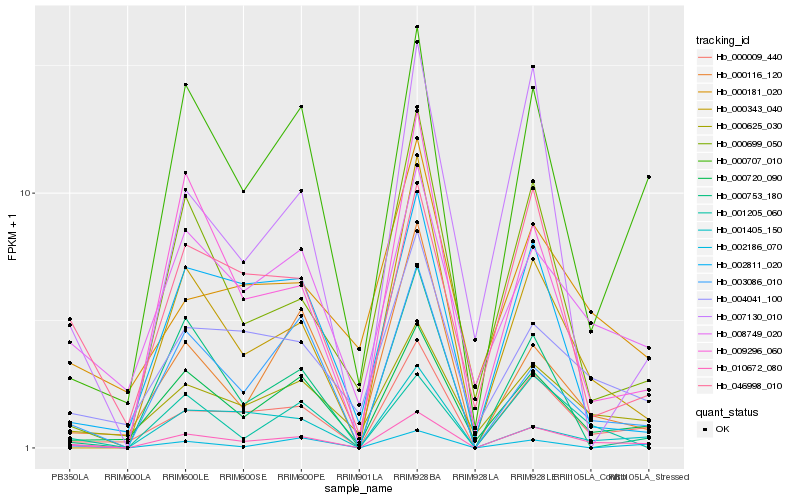
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010672_080 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_00902 [Jatropha curcas] |
| 2 |
Hb_000009_440 |
0.1275385572 |
- |
- |
PREDICTED: uncharacterized protein LOC105639617 [Jatropha curcas] |
| 3 |
Hb_000720_090 |
0.1663051922 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 4 |
Hb_000707_010 |
0.1817262082 |
- |
- |
hypothetical protein JCGZ_25098 [Jatropha curcas] |
| 5 |
Hb_000625_030 |
0.1820209834 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 6 |
Hb_003086_010 |
0.1834595127 |
- |
- |
hypothetical protein JCGZ_06093 [Jatropha curcas] |
| 7 |
Hb_000343_040 |
0.1837769044 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 8 |
Hb_008749_020 |
0.1906632894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004041_100 |
0.1960015519 |
- |
- |
- |
| 10 |
Hb_001205_060 |
0.2023475148 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 11 |
Hb_046998_010 |
0.2052069406 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 12 |
Hb_001405_150 |
0.2067201521 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 13 |
Hb_000699_050 |
0.2093115894 |
- |
- |
- |
| 14 |
Hb_000116_120 |
0.2133962616 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 15 |
Hb_000181_020 |
0.2171194399 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_002811_020 |
0.2175965324 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 17 |
Hb_009296_060 |
0.2218369233 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 18 |
Hb_002186_070 |
0.2232090927 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_000753_180 |
0.2238864342 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 20 |
Hb_007130_010 |
0.2246694907 |
- |
- |
- |