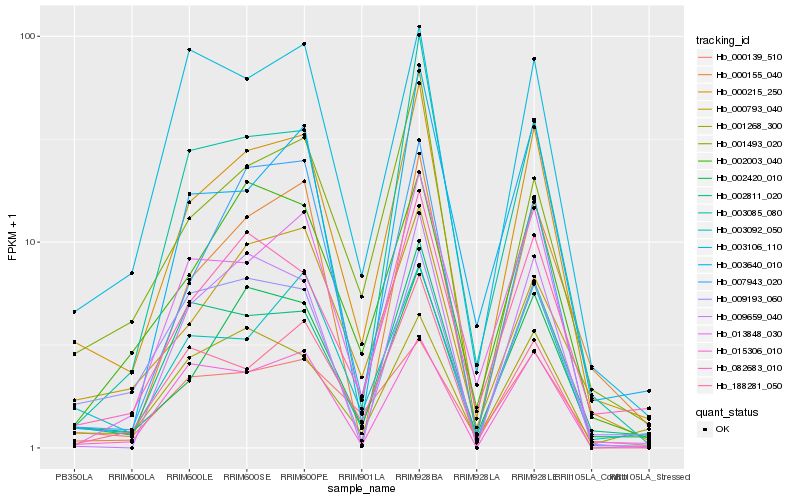
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_250 |
0.0 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_002420_010 |
0.1097808833 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 3 |
Hb_002811_020 |
0.1206189983 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 4 |
Hb_013848_030 |
0.1225157024 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 5 |
Hb_003092_050 |
0.1245735159 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 6 |
Hb_001493_020 |
0.1262696482 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 7 |
Hb_082683_010 |
0.1281101542 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_007943_020 |
0.1313924889 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_002003_040 |
0.1316411238 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003085_080 |
0.1325508646 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_188281_050 |
0.1414954849 |
- |
- |
PREDICTED: solute carrier family 35 member F1-like isoform X1 [Vitis vinifera] |
| 12 |
Hb_000155_040 |
0.1418282831 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 13 |
Hb_015306_010 |
0.1420345112 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 14 |
Hb_009659_040 |
0.145798071 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 15 |
Hb_009193_060 |
0.1464916914 |
- |
- |
PREDICTED: ureide permease 1-like [Jatropha curcas] |
| 16 |
Hb_000139_510 |
0.147735412 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000793_040 |
0.14934371 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 18 |
Hb_001268_300 |
0.1496795736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003640_010 |
0.1503507463 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 20 |
Hb_003106_110 |
0.1512573857 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |