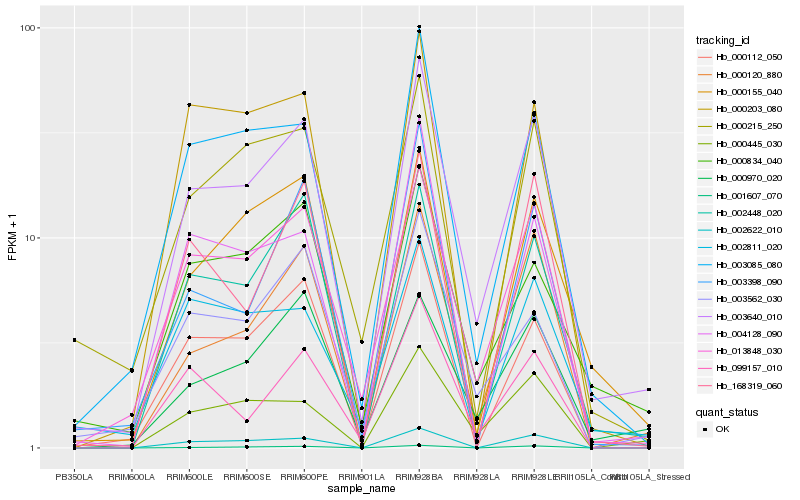
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003640_010 |
0.0 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 2 |
Hb_013848_030 |
0.1017494168 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 3 |
Hb_000120_880 |
0.1114984205 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 4 |
Hb_003085_080 |
0.1186432793 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_000445_030 |
0.1273178328 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 6 |
Hb_002448_020 |
0.1339817244 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 7 |
Hb_002622_010 |
0.1361799576 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 8 |
Hb_000970_020 |
0.1368065279 |
- |
- |
hypothetical protein POPTR_0005s19050g [Populus trichocarpa] |
| 9 |
Hb_000155_040 |
0.1379501599 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 10 |
Hb_000834_040 |
0.1413093751 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 11 |
Hb_000112_050 |
0.1438770816 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 12 |
Hb_003562_030 |
0.1440474812 |
- |
- |
- |
| 13 |
Hb_004128_090 |
0.1472448931 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 14 |
Hb_099157_010 |
0.1497615001 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 15 |
Hb_000215_250 |
0.1503507463 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001607_070 |
0.1522084709 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 17 |
Hb_002811_020 |
0.1530164859 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 18 |
Hb_003398_090 |
0.1532209623 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 19 |
Hb_168319_060 |
0.1544759627 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 20 |
Hb_000203_080 |
0.1556758585 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |