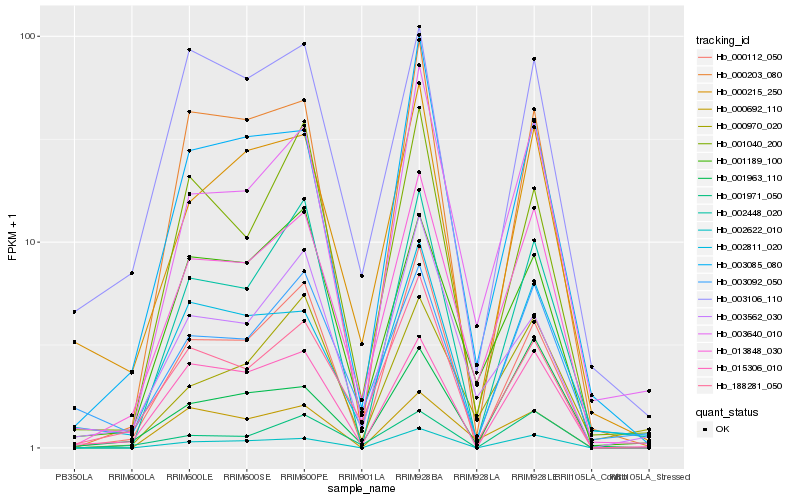
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013848_030 |
0.0 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 2 |
Hb_003640_010 |
0.1017494168 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 3 |
Hb_188281_050 |
0.1145786299 |
- |
- |
PREDICTED: solute carrier family 35 member F1-like isoform X1 [Vitis vinifera] |
| 4 |
Hb_001189_100 |
0.1224719436 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 5 |
Hb_000215_250 |
0.1225157024 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_003085_080 |
0.1286514949 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_002811_020 |
0.136459695 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 8 |
Hb_015306_010 |
0.1380868052 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 9 |
Hb_000203_080 |
0.1434723078 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 10 |
Hb_003092_050 |
0.1442897224 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 11 |
Hb_001971_050 |
0.1448970617 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_002622_010 |
0.1450002653 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 13 |
Hb_000970_020 |
0.1452183123 |
- |
- |
hypothetical protein POPTR_0005s19050g [Populus trichocarpa] |
| 14 |
Hb_000112_050 |
0.1453441447 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 15 |
Hb_003562_030 |
0.1456497205 |
- |
- |
- |
| 16 |
Hb_000692_110 |
0.1461112728 |
- |
- |
PREDICTED: NEP1-interacting protein-like 1 [Jatropha curcas] |
| 17 |
Hb_003106_110 |
0.1473040498 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001963_110 |
0.148747164 |
- |
- |
- |
| 19 |
Hb_001040_200 |
0.1495575252 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 20 |
Hb_002448_020 |
0.1500906005 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |