| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
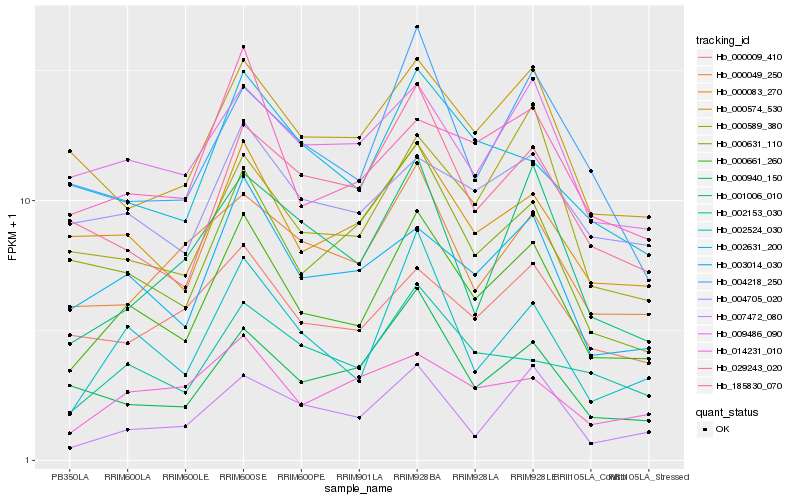
Hb_000661_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 2 |
Hb_002524_030 |
0.1046303889 |
- |
- |
- |
| 3 |
Hb_003014_030 |
0.1053705328 |
- |
- |
PREDICTED: thyroid adenoma-associated protein homolog [Jatropha curcas] |
| 4 |
Hb_000083_270 |
0.1080587681 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 5 |
Hb_029243_020 |
0.1179816986 |
- |
- |
ENTH/VHS/GAT family protein [Theobroma cacao] |
| 6 |
Hb_000631_110 |
0.1196271164 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_000574_530 |
0.1198841111 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 8 |
Hb_002153_030 |
0.1199399027 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 9 |
Hb_009486_090 |
0.1200505363 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000589_380 |
0.1249605221 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 11 |
Hb_185830_070 |
0.1249870799 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000940_150 |
0.1253351872 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 13 |
Hb_000049_250 |
0.1271378752 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000009_410 |
0.127149241 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 15 |
Hb_004218_250 |
0.1280320629 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 16 |
Hb_002631_200 |
0.1284282011 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 17 |
Hb_014231_010 |
0.129612151 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 18 |
Hb_004705_020 |
0.1299228579 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 19 |
Hb_007472_080 |
0.1313778025 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 20 |
Hb_001006_010 |
0.1323505618 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |