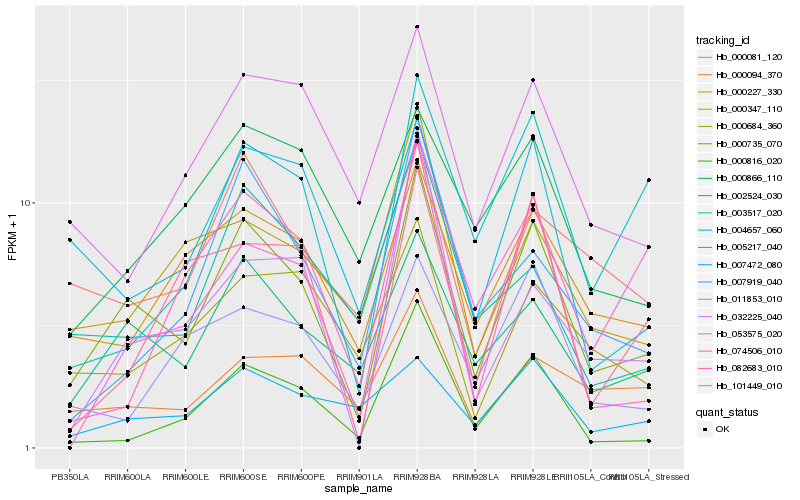
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000735_070 |
0.0 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 2 |
Hb_002524_030 |
0.1433763696 |
- |
- |
- |
| 3 |
Hb_000227_330 |
0.1754693297 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 4 |
Hb_000347_110 |
0.1834139453 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 5 |
Hb_005217_040 |
0.1852156327 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000816_020 |
0.1877034746 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_003517_020 |
0.1885120521 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 8 |
Hb_007919_040 |
0.1898729095 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 9 |
Hb_000081_120 |
0.1901680962 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 10 |
Hb_000094_370 |
0.1909441181 |
- |
- |
- |
| 11 |
Hb_007472_080 |
0.1935445139 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 12 |
Hb_053575_020 |
0.195367635 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 13 |
Hb_004657_060 |
0.1955282295 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 14 |
Hb_000684_360 |
0.1968189277 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 15 |
Hb_011853_010 |
0.1976668626 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 16 |
Hb_032225_040 |
0.19771373 |
- |
- |
- |
| 17 |
Hb_101449_010 |
0.197851786 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 18 |
Hb_000866_110 |
0.2003185222 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 19 |
Hb_082683_010 |
0.2020701556 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_074506_010 |
0.2036999219 |
- |
- |
JHL25H03.10 [Jatropha curcas] |