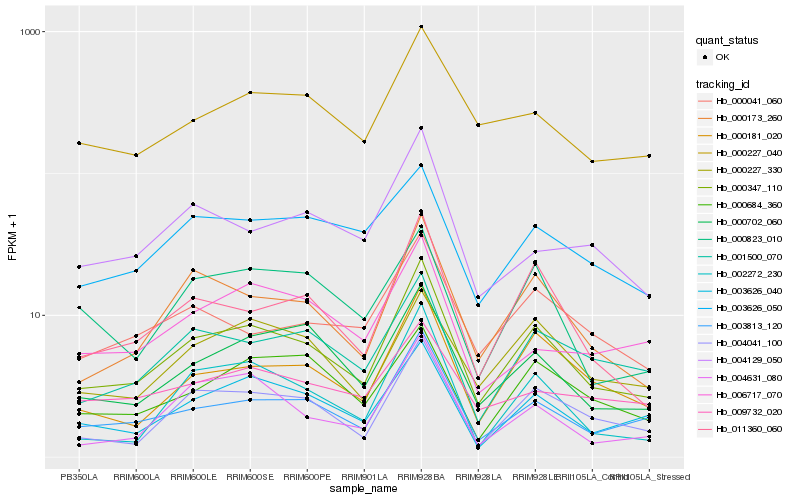
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_110 |
0.0 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 2 |
Hb_003813_120 |
0.0951156154 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000173_260 |
0.0971328917 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 4 |
Hb_004041_100 |
0.1031217331 |
- |
- |
- |
| 5 |
Hb_011360_060 |
0.1086355848 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 6 |
Hb_000181_020 |
0.1119438343 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_003626_040 |
0.111991104 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000702_060 |
0.1180395838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001500_070 |
0.1255462366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002272_230 |
0.1354662663 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 11 |
Hb_000041_060 |
0.1361027916 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 12 |
Hb_000227_040 |
0.1363059618 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 13 |
Hb_000227_330 |
0.1364346706 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 14 |
Hb_004631_080 |
0.1419101117 |
- |
- |
P3B-ATPase PH1 [Petunia x hybrida] |
| 15 |
Hb_000684_360 |
0.1449944073 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 16 |
Hb_004129_050 |
0.1459548222 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 17 |
Hb_003626_050 |
0.1476683682 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 18 |
Hb_000823_010 |
0.1480931118 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 19 |
Hb_006717_070 |
0.1485649058 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 20 |
Hb_009732_020 |
0.1498963393 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |