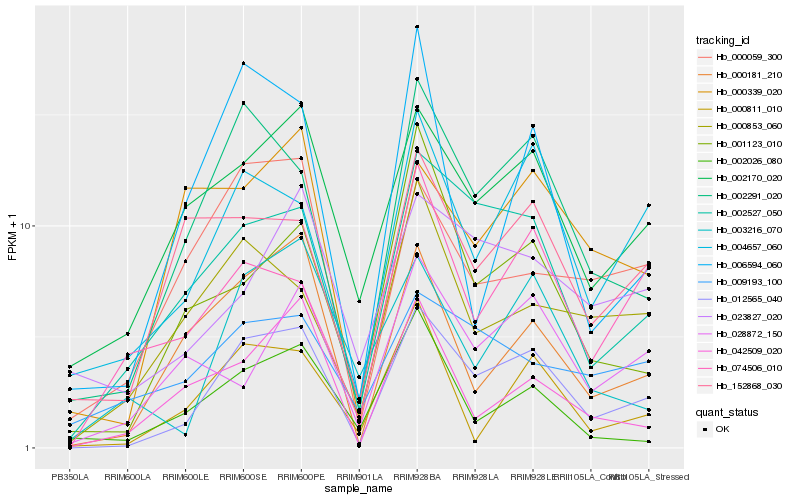
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012565_040 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 2 |
Hb_002291_020 |
0.1596289563 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002527_050 |
0.1604060347 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 4 |
Hb_002170_020 |
0.1778636285 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 5 |
Hb_004657_060 |
0.1850736975 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 6 |
Hb_000181_210 |
0.190638587 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_000059_300 |
0.2100238287 |
- |
- |
caspase, putative [Ricinus communis] |
| 8 |
Hb_000811_010 |
0.2125822751 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 9 |
Hb_003216_070 |
0.2162452109 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 10 |
Hb_152868_030 |
0.216766771 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 11 |
Hb_001123_010 |
0.2201394544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000853_060 |
0.2211889159 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_074506_010 |
0.2225133074 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 14 |
Hb_006594_060 |
0.2236689909 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 15 |
Hb_042509_020 |
0.2236740528 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 16 |
Hb_023827_020 |
0.2238238472 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 17 |
Hb_000339_020 |
0.2247082808 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 18 |
Hb_028872_150 |
0.2252816406 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |
| 19 |
Hb_002026_080 |
0.230716676 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 20 |
Hb_009193_100 |
0.230875861 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |