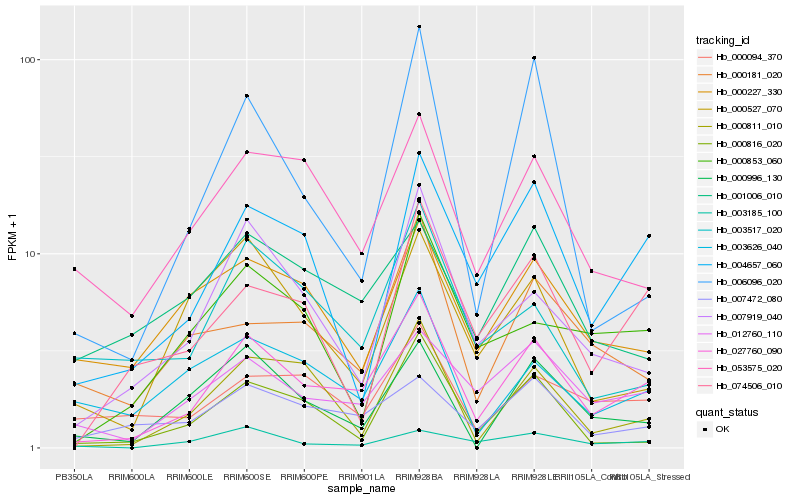
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027760_090 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_012760_110 |
0.1691391378 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004657_060 |
0.180089833 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 4 |
Hb_007919_040 |
0.1886153535 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 5 |
Hb_007472_080 |
0.1912413948 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 6 |
Hb_000811_010 |
0.1922050055 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 7 |
Hb_074506_010 |
0.2000726129 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 8 |
Hb_053575_020 |
0.2007763187 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 9 |
Hb_003626_040 |
0.2010080385 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000094_370 |
0.2010922666 |
- |
- |
- |
| 11 |
Hb_000181_020 |
0.2052254198 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000996_130 |
0.2079771769 |
- |
- |
PREDICTED: remorin-like [Populus euphratica] |
| 13 |
Hb_003185_100 |
0.2084613325 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 14 |
Hb_000816_020 |
0.2123841036 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_000853_060 |
0.2133956242 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000227_330 |
0.2141535054 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 17 |
Hb_006096_020 |
0.215992132 |
- |
- |
Os06g0343500 [Oryza sativa Japonica Group] |
| 18 |
Hb_000527_070 |
0.2165508394 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 19 |
Hb_001006_010 |
0.2178539458 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 20 |
Hb_003517_020 |
0.2200708934 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |