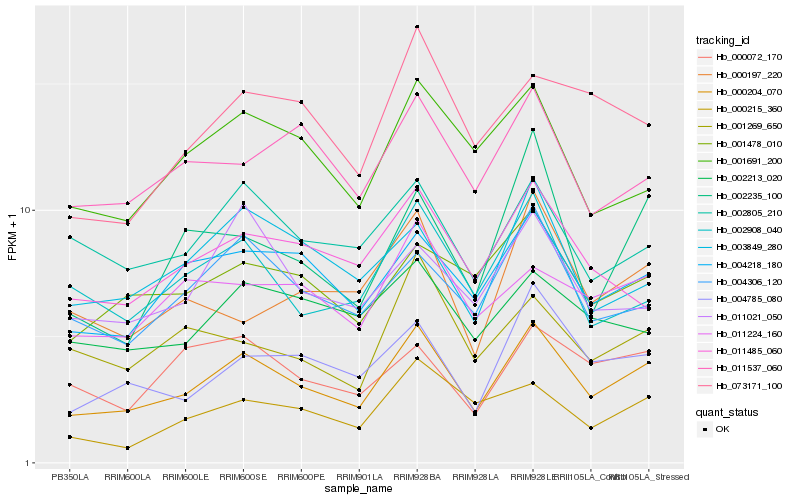
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000204_070 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 2 |
Hb_004218_180 |
0.107647947 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 3 |
Hb_004785_080 |
0.1091846625 |
- |
- |
hypothetical protein POPTR_0005s21250g [Populus trichocarpa] |
| 4 |
Hb_004306_120 |
0.1124383227 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002908_040 |
0.1185745575 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 6 |
Hb_003849_280 |
0.120777479 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 7 |
Hb_001478_010 |
0.1220596625 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48250, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002805_210 |
0.1231431374 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 9 |
Hb_001691_200 |
0.1239532326 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 10 |
Hb_002235_100 |
0.125149013 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011224_160 |
0.1266482041 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 12 |
Hb_011537_060 |
0.1271190652 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 13 |
Hb_000215_360 |
0.1275767752 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_073171_100 |
0.1290151721 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 15 |
Hb_002213_020 |
0.1296209569 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011485_060 |
0.1306820818 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 17 |
Hb_000197_220 |
0.1309452403 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000072_170 |
0.1317815485 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 19 |
Hb_011021_050 |
0.1318821844 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001269_650 |
0.1320645424 |
- |
- |
transporter, putative [Ricinus communis] |