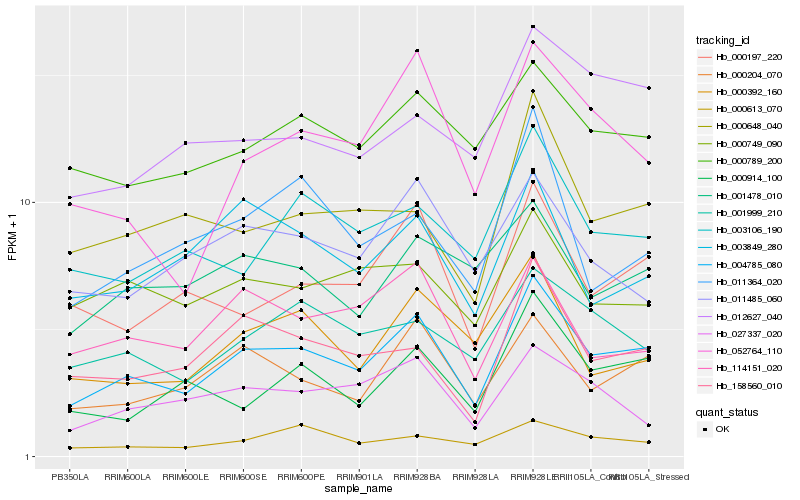
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004785_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s21250g [Populus trichocarpa] |
| 2 |
Hb_000204_070 |
0.1091846625 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 3 |
Hb_000197_220 |
0.1170491897 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_158560_010 |
0.1236405517 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 5 |
Hb_052764_110 |
0.1259999164 |
- |
- |
PREDICTED: quinone oxidoreductase PIG3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001999_210 |
0.1281942437 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic [Eucalyptus grandis] |
| 7 |
Hb_000789_200 |
0.1282622989 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 8 |
Hb_000648_040 |
0.13021096 |
- |
- |
unknown [Populus trichocarpa] |
| 9 |
Hb_114151_020 |
0.1307373054 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 10 |
Hb_003106_190 |
0.1310676256 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 11 |
Hb_000392_160 |
0.1333431454 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000613_070 |
0.1345763046 |
- |
- |
- |
| 13 |
Hb_000914_100 |
0.1353965979 |
- |
- |
unknown [Glycine max] |
| 14 |
Hb_001478_010 |
0.1362737494 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48250, chloroplastic [Jatropha curcas] |
| 15 |
Hb_027337_020 |
0.1369701928 |
- |
- |
sodium/hydrogen exchanger plant, putative [Ricinus communis] |
| 16 |
Hb_000749_090 |
0.1380293265 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_003849_280 |
0.1384561129 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 18 |
Hb_012627_040 |
0.1389072476 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 19 |
Hb_011364_020 |
0.1393985761 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 20 |
Hb_011485_060 |
0.1396222446 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |