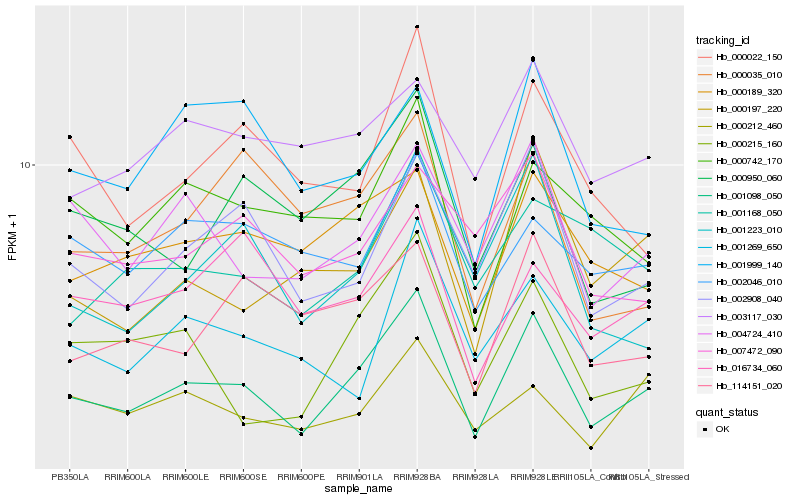
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001098_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 2 |
Hb_002908_040 |
0.1179303234 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 3 |
Hb_004724_410 |
0.1248158642 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000197_220 |
0.1297453206 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001223_010 |
0.1298457559 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 6 |
Hb_000215_160 |
0.1301166779 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000950_060 |
0.1349331475 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 8 |
Hb_000189_320 |
0.1358700838 |
- |
- |
PREDICTED: multisubstrate pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000022_150 |
0.1361030642 |
- |
- |
- |
| 10 |
Hb_114151_020 |
0.1370519533 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 11 |
Hb_001269_650 |
0.1416333814 |
- |
- |
transporter, putative [Ricinus communis] |
| 12 |
Hb_000742_170 |
0.142090502 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_000212_460 |
0.1434187366 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 14 |
Hb_016734_060 |
0.1463764543 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 15 |
Hb_001999_140 |
0.1482610345 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_003117_030 |
0.1493908686 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 17 |
Hb_001168_050 |
0.149596036 |
- |
- |
hypothetical protein POPTR_0007s03190g [Populus trichocarpa] |
| 18 |
Hb_007472_090 |
0.1498943928 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_000035_010 |
0.1509938496 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002046_010 |
0.1510349519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |