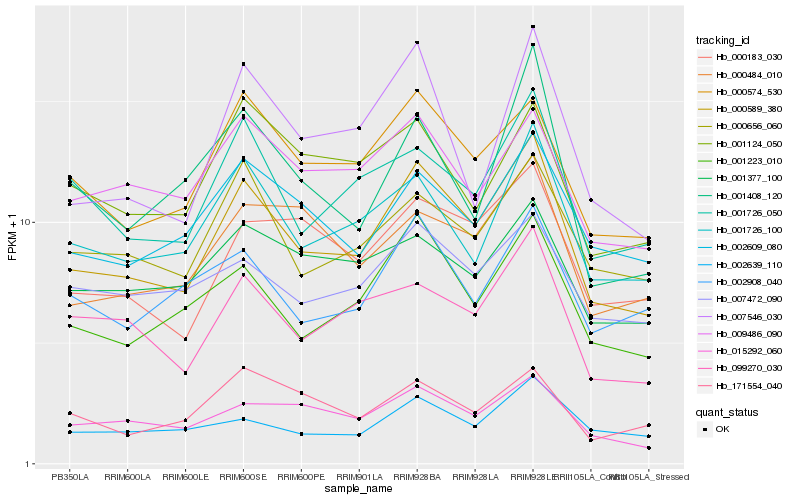
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000589_380 |
0.0 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 2 |
Hb_001223_010 |
0.0899241184 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 3 |
Hb_001726_100 |
0.0936465015 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000574_530 |
0.0939669334 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 5 |
Hb_099270_030 |
0.1025962125 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 6 |
Hb_015292_060 |
0.1027737706 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 7 |
Hb_001726_050 |
0.1031657804 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 8 |
Hb_000656_060 |
0.103267776 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_002609_080 |
0.1053551231 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000183_030 |
0.1056964652 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0291141 [Populus euphratica] |
| 11 |
Hb_007546_030 |
0.109703034 |
- |
- |
transcription cofactor, putative [Ricinus communis] |
| 12 |
Hb_002639_110 |
0.1124690015 |
- |
- |
hypothetical protein L484_011673 [Morus notabilis] |
| 13 |
Hb_000484_010 |
0.1127117545 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 14 |
Hb_009486_090 |
0.1128669678 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_002908_040 |
0.1154858847 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 16 |
Hb_007472_090 |
0.1177361546 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_001377_100 |
0.1186866342 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001408_120 |
0.120460517 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_171554_040 |
0.1212340921 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 20 |
Hb_001124_050 |
0.1217008143 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |