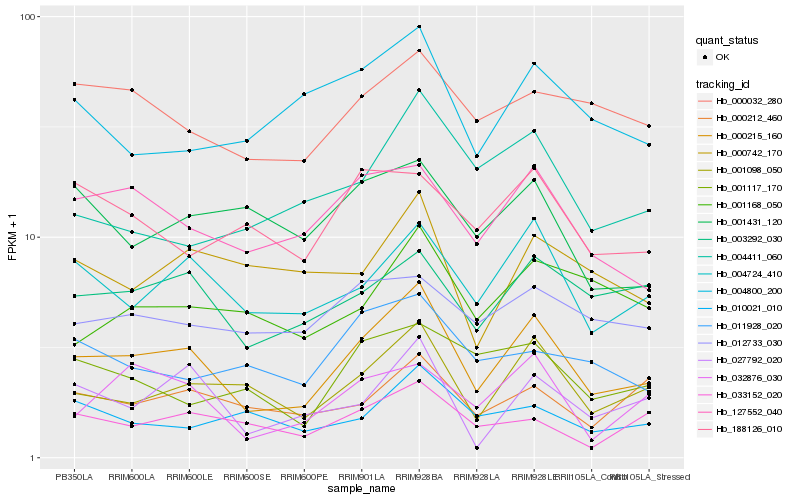
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_160 |
0.0 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001098_050 |
0.1301166779 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 3 |
Hb_004724_410 |
0.1343617939 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_127552_040 |
0.1507691294 |
- |
- |
Uncharacterized protein L484_000643 [Morus notabilis] |
| 5 |
Hb_003292_030 |
0.1538769867 |
- |
- |
PREDICTED: exosome complex component RRP42 [Populus euphratica] |
| 6 |
Hb_027792_020 |
0.1566002311 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 7 |
Hb_032876_030 |
0.1633769884 |
- |
- |
- |
| 8 |
Hb_000032_280 |
0.1674648719 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 9 |
Hb_000212_460 |
0.1701134312 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 10 |
Hb_033152_020 |
0.1707674063 |
- |
- |
hypothetical protein VITISV_000649 [Vitis vinifera] |
| 11 |
Hb_004800_200 |
0.1748378901 |
- |
- |
- |
| 12 |
Hb_001117_170 |
0.1750686574 |
- |
- |
- |
| 13 |
Hb_000742_170 |
0.1758095173 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_004411_060 |
0.1768048077 |
- |
- |
acetolactate synthase, putative [Ricinus communis] |
| 15 |
Hb_012733_030 |
0.1769863031 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |
| 16 |
Hb_001168_050 |
0.1777577678 |
- |
- |
hypothetical protein POPTR_0007s03190g [Populus trichocarpa] |
| 17 |
Hb_011928_020 |
0.1788469368 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |
| 18 |
Hb_188126_010 |
0.1795948131 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_010021_010 |
0.1797507642 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_001431_120 |
0.1799872189 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |