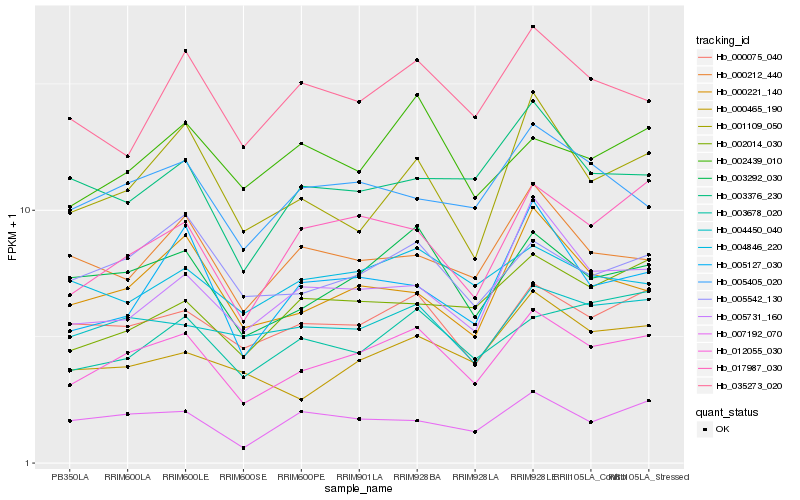
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012055_030 |
0.0 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 2 |
Hb_003292_030 |
0.0689988097 |
- |
- |
PREDICTED: exosome complex component RRP42 [Populus euphratica] |
| 3 |
Hb_000075_040 |
0.0836713819 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 4 |
Hb_017987_030 |
0.0896632035 |
- |
- |
PREDICTED: uncharacterized protein LOC105630796 [Jatropha curcas] |
| 5 |
Hb_005542_130 |
0.0956668382 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 6 |
Hb_004450_040 |
0.0958141917 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |
| 7 |
Hb_001109_050 |
0.1019739446 |
- |
- |
PREDICTED: ankyrin-1 [Jatropha curcas] |
| 8 |
Hb_003678_020 |
0.1032978604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005405_020 |
0.1033714746 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 10 |
Hb_007192_070 |
0.1038392813 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_002014_030 |
0.1041461683 |
- |
- |
hypothetical protein JCGZ_07251 [Jatropha curcas] |
| 12 |
Hb_000221_140 |
0.1044400928 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_002439_010 |
0.1050432311 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 14 |
Hb_035273_020 |
0.1054885379 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_005731_160 |
0.1059389604 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_000465_190 |
0.1091298015 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000212_440 |
0.1095460547 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_003376_230 |
0.1100871461 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 19 |
Hb_005127_030 |
0.1101891109 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 20 |
Hb_004846_220 |
0.1102336474 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |