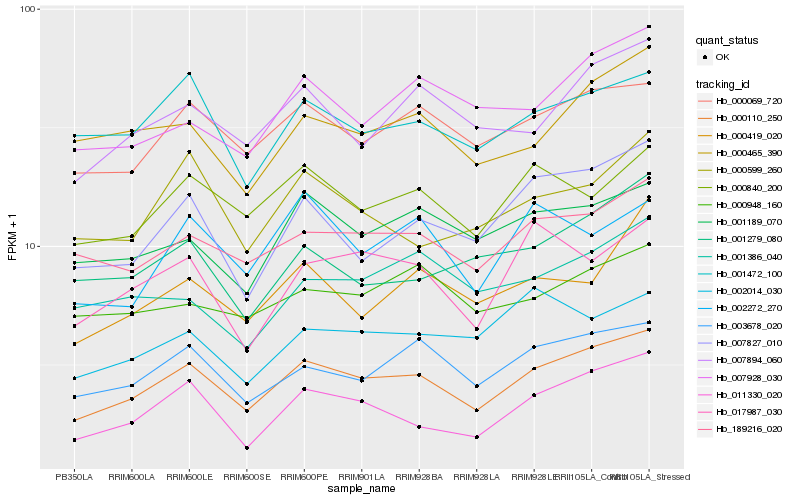
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_250 |
0.0 |
- |
- |
PREDICTED: homologous-pairing protein 2 homolog [Jatropha curcas] |
| 2 |
Hb_000069_720 |
0.0730806644 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |
| 3 |
Hb_003678_020 |
0.0736601175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_189216_020 |
0.0809347124 |
- |
- |
PREDICTED: translin [Jatropha curcas] |
| 5 |
Hb_011330_020 |
0.0818447926 |
transcription factor |
TF Family: ARID |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_007894_060 |
0.0837811126 |
- |
- |
PREDICTED: ubiquitin thioesterase OTU1 [Jatropha curcas] |
| 7 |
Hb_007827_010 |
0.0851701009 |
- |
- |
PREDICTED: uncharacterized protein C12B10.15c [Jatropha curcas] |
| 8 |
Hb_000840_200 |
0.0875152685 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_001472_100 |
0.0886300815 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 10 |
Hb_001189_070 |
0.0894278368 |
- |
- |
PREDICTED: ribosome production factor 1 [Jatropha curcas] |
| 11 |
Hb_000465_390 |
0.0916624339 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Jatropha curcas] |
| 12 |
Hb_007928_030 |
0.0932142721 |
- |
- |
PREDICTED: dual specificity phosphatase Cdc25 [Jatropha curcas] |
| 13 |
Hb_017987_030 |
0.0945878225 |
- |
- |
PREDICTED: uncharacterized protein LOC105630796 [Jatropha curcas] |
| 14 |
Hb_000599_260 |
0.0965895263 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 15 |
Hb_000948_160 |
0.0992235323 |
- |
- |
PREDICTED: 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000419_020 |
0.1004993172 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_001279_080 |
0.10126095 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105140180 isoform X1 [Populus euphratica] |
| 18 |
Hb_002272_270 |
0.1014037405 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 19 |
Hb_002014_030 |
0.1016507984 |
- |
- |
hypothetical protein JCGZ_07251 [Jatropha curcas] |
| 20 |
Hb_001386_040 |
0.1020933337 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 511 [Jatropha curcas] |