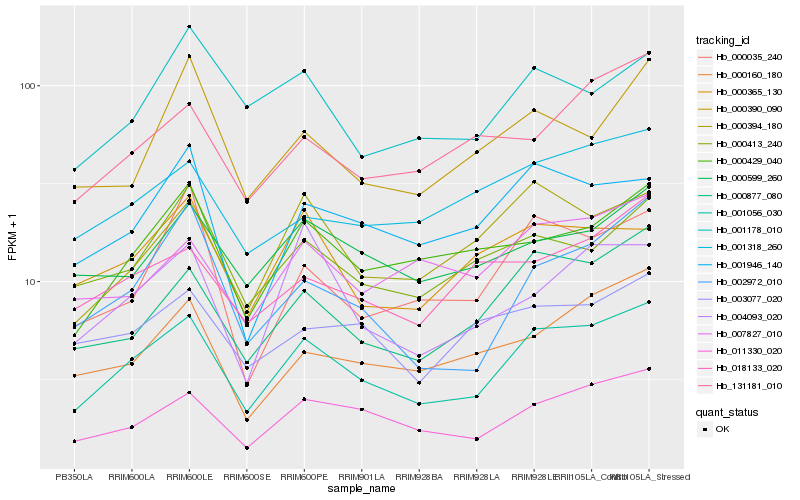
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001056_030 |
0.0 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |
| 2 |
Hb_011330_020 |
0.0881202513 |
transcription factor |
TF Family: ARID |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000877_080 |
0.092001772 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |
| 4 |
Hb_000429_040 |
0.1140200724 |
- |
- |
PREDICTED: chaperone protein dnaJ 6 [Vitis vinifera] |
| 5 |
Hb_000394_180 |
0.116094442 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 6 |
Hb_002972_010 |
0.1172149985 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 7 |
Hb_000413_240 |
0.1187022607 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000599_260 |
0.118871902 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 9 |
Hb_000160_180 |
0.1203478722 |
- |
- |
methionyl-tRNA formyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001946_140 |
0.1218326386 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_001318_260 |
0.1236085948 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 12 |
Hb_007827_010 |
0.1236331461 |
- |
- |
PREDICTED: uncharacterized protein C12B10.15c [Jatropha curcas] |
| 13 |
Hb_000035_240 |
0.1262725125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_131181_010 |
0.127611553 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |
| 15 |
Hb_004093_020 |
0.1282050999 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |
| 16 |
Hb_018133_020 |
0.1290071692 |
- |
- |
transporter, putative [Ricinus communis] |
| 17 |
Hb_003077_020 |
0.1291680679 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000365_130 |
0.1296105128 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 19 |
Hb_001178_010 |
0.1309027341 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 20 |
Hb_000390_090 |
0.1328284624 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |