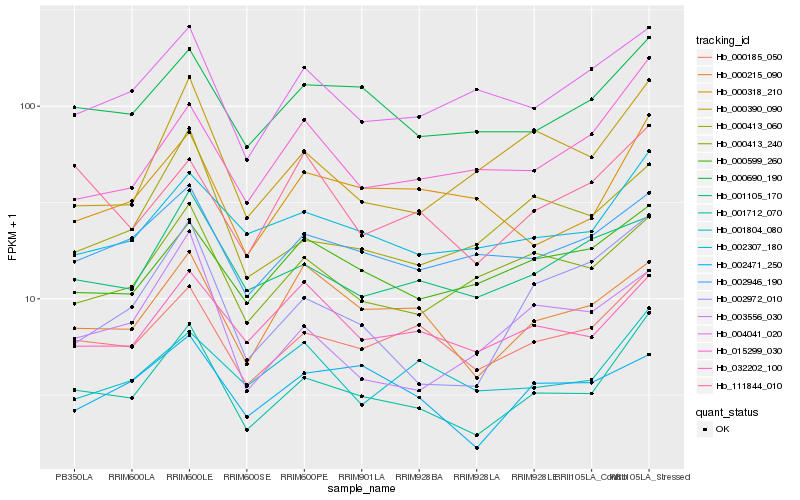
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001712_070 |
0.0 |
- |
- |
PREDICTED: HD domain-containing protein 2 [Jatropha curcas] |
| 2 |
Hb_000185_050 |
0.1012805901 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 3 |
Hb_000690_190 |
0.1120474596 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 4 |
Hb_002307_180 |
0.1160725206 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_015299_030 |
0.12702557 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 6 |
Hb_000215_090 |
0.1270721309 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_000390_090 |
0.1272192769 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 8 |
Hb_002946_190 |
0.1288748535 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000318_210 |
0.129303637 |
- |
- |
PREDICTED: 40S ribosomal protein S12-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002972_010 |
0.1297186657 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 11 |
Hb_001105_170 |
0.1297908639 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000413_240 |
0.1312359694 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000413_060 |
0.1338317621 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 14 |
Hb_004041_020 |
0.1339794152 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 15 |
Hb_000599_260 |
0.1351205834 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 16 |
Hb_111844_010 |
0.1363219846 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 17 |
Hb_032202_100 |
0.1388584254 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 18 |
Hb_001804_080 |
0.1438846002 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 19 |
Hb_002471_250 |
0.1457324427 |
- |
- |
Tapt1/CMV receptor, putative isoform 2 [Theobroma cacao] |
| 20 |
Hb_003556_030 |
0.1469093438 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |