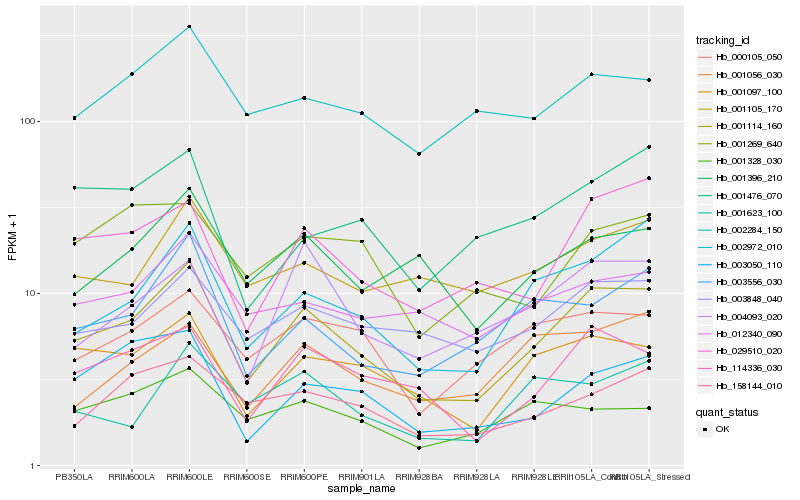
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001114_160 |
0.0 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002972_010 |
0.1233905828 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 3 |
Hb_114336_030 |
0.1246723545 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 4 |
Hb_001097_100 |
0.1268607394 |
- |
- |
Chaperone DnaJ-domain superfamily protein [Theobroma cacao] |
| 5 |
Hb_029510_020 |
0.1298053161 |
- |
- |
hypothetical protein JCGZ_06306 [Jatropha curcas] |
| 6 |
Hb_003848_040 |
0.1392317528 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_003556_030 |
0.1408332078 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 8 |
Hb_158144_010 |
0.1450063942 |
- |
- |
- |
| 9 |
Hb_000105_050 |
0.1458104152 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 10 |
Hb_001396_210 |
0.1480881349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003050_110 |
0.1493679915 |
- |
- |
PREDICTED: prefoldin subunit 6 [Populus euphratica] |
| 12 |
Hb_012340_090 |
0.1526272037 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 13 |
Hb_004093_020 |
0.1528811413 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |
| 14 |
Hb_002284_150 |
0.1542635189 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 15 |
Hb_001105_170 |
0.1547371822 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001476_070 |
0.1563599328 |
- |
- |
MinE protein [Manihot esculenta] |
| 17 |
Hb_001056_030 |
0.1569535929 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001623_100 |
0.1613546336 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 19 |
Hb_001328_030 |
0.1618675973 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 20 |
Hb_001269_640 |
0.1628638445 |
- |
- |
hypothetical protein CISIN_1g032720mg [Citrus sinensis] |