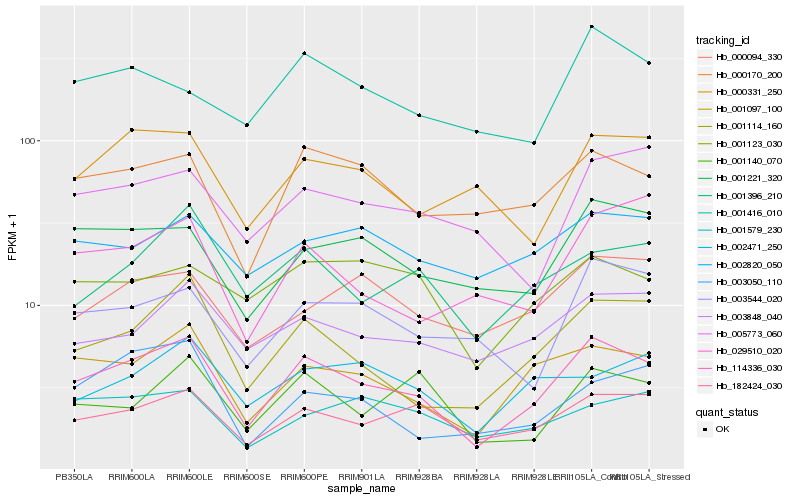
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114336_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 2 |
Hb_001097_100 |
0.1147179073 |
- |
- |
Chaperone DnaJ-domain superfamily protein [Theobroma cacao] |
| 3 |
Hb_001114_160 |
0.1246723545 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 4 |
Hb_182424_030 |
0.1288868027 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 8 [Vitis vinifera] |
| 5 |
Hb_000170_200 |
0.1408101325 |
- |
- |
PREDICTED: protein KRTCAP2 homolog [Jatropha curcas] |
| 6 |
Hb_001221_320 |
0.1425094542 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003848_040 |
0.1481679026 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_001396_210 |
0.1484631863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003544_020 |
0.1547036129 |
- |
- |
PREDICTED: elongin-A [Jatropha curcas] |
| 10 |
Hb_000331_250 |
0.1559528008 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Vitis vinifera] |
| 11 |
Hb_001140_070 |
0.1573154551 |
- |
- |
Chromosome transmission fidelity protein 8, putative isoform 2 [Theobroma cacao] |
| 12 |
Hb_001416_010 |
0.1593440889 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 13 |
Hb_000094_330 |
0.1607252733 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 4 [Jatropha curcas] |
| 14 |
Hb_001123_030 |
0.160955164 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_002471_250 |
0.1617137106 |
- |
- |
Tapt1/CMV receptor, putative isoform 2 [Theobroma cacao] |
| 16 |
Hb_005773_060 |
0.1628015698 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 17 |
Hb_002820_050 |
0.1638827837 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 18 |
Hb_003050_110 |
0.1640129939 |
- |
- |
PREDICTED: prefoldin subunit 6 [Populus euphratica] |
| 19 |
Hb_029510_020 |
0.1641028375 |
- |
- |
hypothetical protein JCGZ_06306 [Jatropha curcas] |
| 20 |
Hb_001579_230 |
0.1651758025 |
- |
- |
PREDICTED: uncharacterized protein LOC105644968 [Jatropha curcas] |