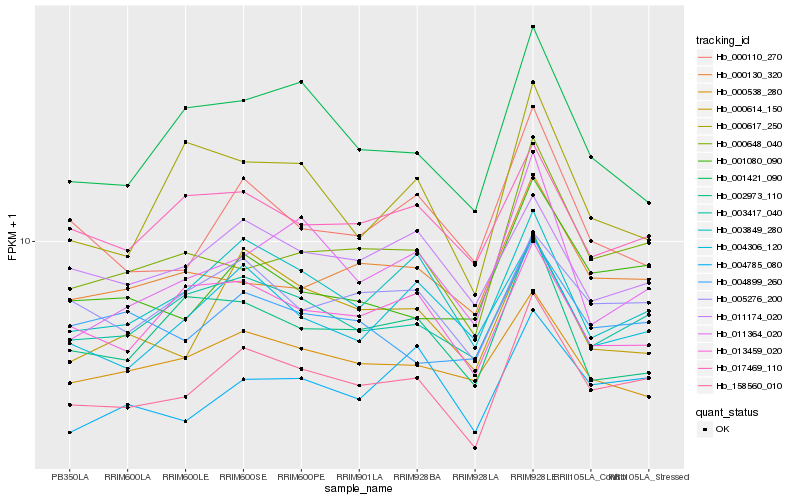
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158560_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 2 |
Hb_000648_040 |
0.1056368608 |
- |
- |
unknown [Populus trichocarpa] |
| 3 |
Hb_003417_040 |
0.1131572097 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 4 |
Hb_003849_280 |
0.1147451995 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 5 |
Hb_000614_150 |
0.1182210213 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 6 |
Hb_001080_090 |
0.1193160029 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000110_270 |
0.1204123596 |
- |
- |
prp4, putative [Ricinus communis] |
| 8 |
Hb_011364_020 |
0.121157781 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005276_200 |
0.1211949837 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004785_080 |
0.1236405517 |
- |
- |
hypothetical protein POPTR_0005s21250g [Populus trichocarpa] |
| 11 |
Hb_001421_090 |
0.1286163544 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 12 |
Hb_013459_020 |
0.1300645988 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 13 |
Hb_000130_320 |
0.1308469822 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 14 |
Hb_000617_250 |
0.1333468723 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004306_120 |
0.1338074345 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_004899_260 |
0.1338331952 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 17 |
Hb_002973_110 |
0.1348741212 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000538_280 |
0.1368617471 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 19 |
Hb_011174_020 |
0.1380916916 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 20 |
Hb_017469_110 |
0.1395051266 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |