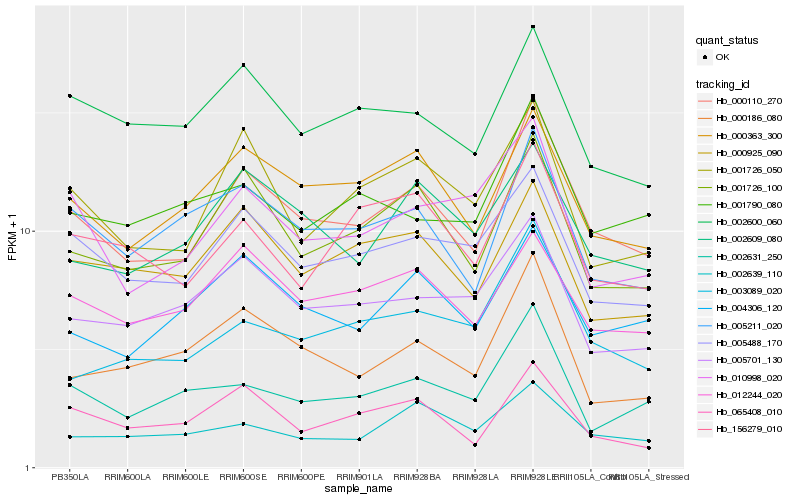
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_270 |
0.0 |
- |
- |
prp4, putative [Ricinus communis] |
| 2 |
Hb_001726_100 |
0.0860379986 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000363_300 |
0.0895976006 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005488_170 |
0.0976590139 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 5 |
Hb_005211_020 |
0.0999538379 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004306_120 |
0.1012882685 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_012244_020 |
0.103885343 |
- |
- |
calpain, putative [Ricinus communis] |
| 8 |
Hb_001726_050 |
0.1042529614 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_002609_080 |
0.106760149 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_002600_060 |
0.1080832138 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 11 |
Hb_005701_130 |
0.1099249679 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 12 |
Hb_002639_110 |
0.1102622959 |
- |
- |
hypothetical protein L484_011673 [Morus notabilis] |
| 13 |
Hb_000925_090 |
0.1113047881 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002631_250 |
0.1117553437 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g20730 [Jatropha curcas] |
| 15 |
Hb_000186_080 |
0.1122613442 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 16 |
Hb_156279_010 |
0.112377845 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003089_020 |
0.1129294748 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g18940, chloroplastic [Jatropha curcas] |
| 18 |
Hb_065408_010 |
0.1136760718 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001790_080 |
0.113722546 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 20 |
Hb_010998_020 |
0.1139284955 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |