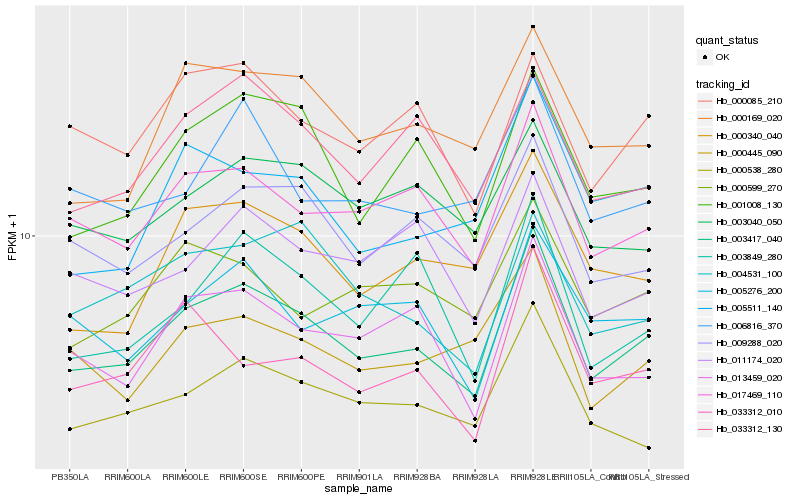
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003417_040 |
0.0 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 2 |
Hb_017469_110 |
0.0676736606 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 3 |
Hb_000538_280 |
0.0763301059 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 4 |
Hb_001008_130 |
0.0773911723 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 5 |
Hb_009288_020 |
0.0798306066 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 6 |
Hb_013459_020 |
0.0813814862 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 7 |
Hb_000169_020 |
0.0841089255 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 8 |
Hb_000340_040 |
0.0847618224 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 9 |
Hb_033312_130 |
0.0850059976 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000085_210 |
0.0884842161 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 11 |
Hb_003040_050 |
0.0895410688 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 12 |
Hb_005511_140 |
0.0898672644 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 13 |
Hb_003849_280 |
0.0899491131 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 14 |
Hb_005276_200 |
0.0905479967 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000445_090 |
0.0906861983 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 16 |
Hb_000599_270 |
0.0907884785 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 17 |
Hb_006816_370 |
0.0910553019 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 18 |
Hb_011174_020 |
0.0916071318 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 19 |
Hb_004531_100 |
0.0916092148 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_033312_010 |
0.0918730337 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |