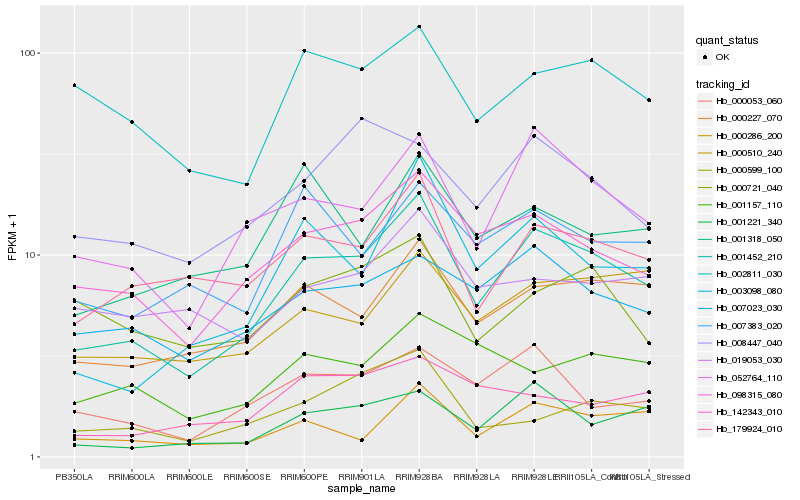
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001452_210 |
0.0 |
- |
- |
molybdopterin biosynthesis CNX1 protein [Arabidopsis thaliana] |
| 2 |
Hb_052764_110 |
0.1110030593 |
- |
- |
PREDICTED: quinone oxidoreductase PIG3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_098315_080 |
0.1136601959 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000227_070 |
0.1205291871 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 5 |
Hb_179924_010 |
0.1288181819 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_003098_080 |
0.1345900203 |
- |
- |
- |
| 7 |
Hb_001221_340 |
0.1391204132 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 8 |
Hb_000053_060 |
0.1401026975 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 9 |
Hb_007023_030 |
0.1411798991 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20710, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_000286_200 |
0.1433208333 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000510_240 |
0.1456552029 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_007383_020 |
0.1491638081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001318_050 |
0.1555430322 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 14 |
Hb_002811_030 |
0.156796398 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 15 |
Hb_008447_040 |
0.1583890324 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 16 |
Hb_142343_010 |
0.1612826256 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_001157_110 |
0.1634734544 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 18 |
Hb_019053_030 |
0.1636062124 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 19 |
Hb_000599_100 |
0.1652371956 |
- |
- |
hypothetical protein B456_001G254000 [Gossypium raimondii] |
| 20 |
Hb_000721_040 |
0.1660924017 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |