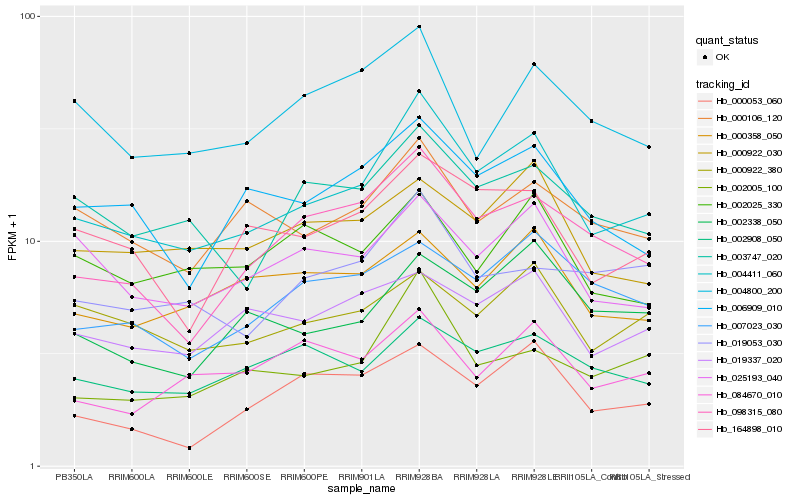
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004411_060 |
0.0 |
- |
- |
acetolactate synthase, putative [Ricinus communis] |
| 2 |
Hb_098315_080 |
0.1046165179 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_164898_010 |
0.108838234 |
- |
- |
Cysteine-rich protein, putative [Theobroma cacao] |
| 4 |
Hb_006909_010 |
0.1098986404 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 5 |
Hb_002005_100 |
0.1106968358 |
- |
- |
- |
| 6 |
Hb_025193_040 |
0.1134584217 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 7 |
Hb_003747_020 |
0.114582837 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X1 [Jatropha curcas] |
| 8 |
Hb_000106_120 |
0.1197478159 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 9 |
Hb_002338_050 |
0.1202297192 |
- |
- |
PREDICTED: uncharacterized protein LOC105118489 isoform X1 [Populus euphratica] |
| 10 |
Hb_019337_020 |
0.1207999128 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 11 |
Hb_002908_050 |
0.1218344086 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 12 |
Hb_000922_030 |
0.1225128345 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_007023_030 |
0.1227709128 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20710, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_019053_030 |
0.1233251878 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 15 |
Hb_000358_050 |
0.1241221317 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 16 |
Hb_000922_380 |
0.1247885934 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein MPP10 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000053_060 |
0.1280117086 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 18 |
Hb_004800_200 |
0.1304103147 |
- |
- |
- |
| 19 |
Hb_084670_010 |
0.130807648 |
- |
- |
PREDICTED: thiamine biosynthetic bifunctional enzyme TH1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_002025_330 |
0.1319394102 |
- |
- |
peptide transporter, putative [Ricinus communis] |