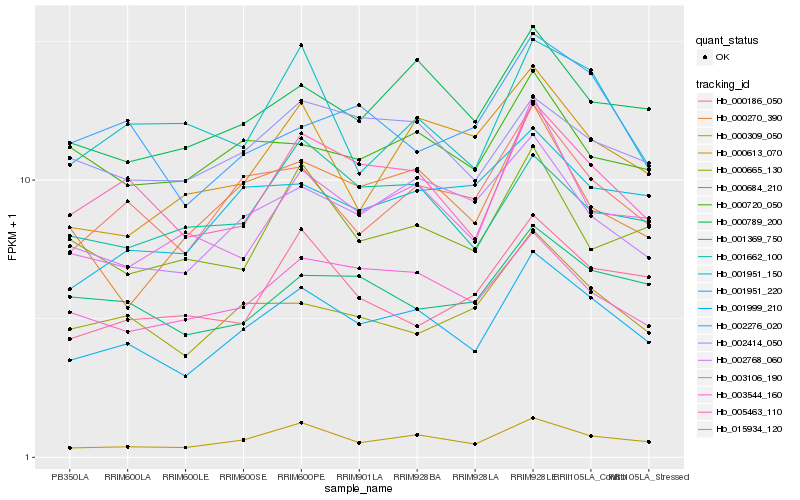
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_210 |
0.0 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic [Eucalyptus grandis] |
| 2 |
Hb_005463_110 |
0.0717716515 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 3 |
Hb_000613_070 |
0.071962658 |
- |
- |
- |
| 4 |
Hb_000186_050 |
0.0785799393 |
- |
- |
PREDICTED: uncharacterized protein LOC105649281 [Jatropha curcas] |
| 5 |
Hb_000789_200 |
0.0887734277 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 6 |
Hb_003544_160 |
0.0899543744 |
- |
- |
- |
| 7 |
Hb_002768_060 |
0.0905569685 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000665_130 |
0.0979481197 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002414_050 |
0.0995115984 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 10 |
Hb_003106_190 |
0.1013579965 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 11 |
Hb_001951_220 |
0.1028228256 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 12 |
Hb_001369_750 |
0.1039847635 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 13 |
Hb_001662_100 |
0.1043759313 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 14 |
Hb_000720_050 |
0.1090219161 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002276_020 |
0.1094502236 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000270_390 |
0.1100861189 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 17 |
Hb_001951_150 |
0.1105395996 |
- |
- |
PREDICTED: uncharacterized protein LOC105649758 [Jatropha curcas] |
| 18 |
Hb_000684_210 |
0.1126007889 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 19 |
Hb_000309_050 |
0.1136052506 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_015934_120 |
0.1144190785 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |