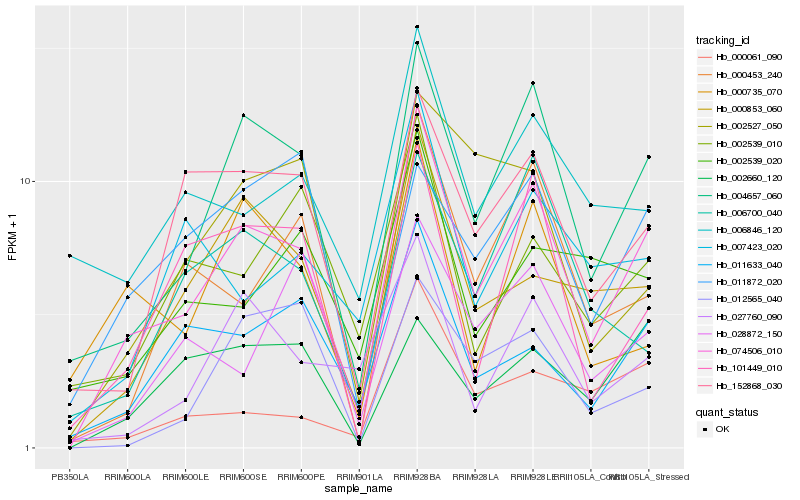
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074506_010 |
0.0 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 2 |
Hb_004657_060 |
0.1168241445 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 3 |
Hb_000853_060 |
0.1800224939 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000453_240 |
0.1862845017 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 5 |
Hb_152868_030 |
0.1869388603 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 6 |
Hb_011633_040 |
0.1928684174 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 7 |
Hb_101449_010 |
0.1972400363 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 8 |
Hb_027760_090 |
0.2000726129 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000735_070 |
0.2036999219 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 10 |
Hb_006700_040 |
0.2079490371 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 11 |
Hb_000061_090 |
0.2099317625 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 12 |
Hb_006846_120 |
0.2102171866 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 13 |
Hb_028872_150 |
0.2104912603 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |
| 14 |
Hb_002660_120 |
0.2116264373 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 15 |
Hb_007423_020 |
0.2123633183 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 16 |
Hb_002539_010 |
0.2143608892 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 17 |
Hb_002527_050 |
0.216206678 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 18 |
Hb_011872_020 |
0.2189591679 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 19 |
Hb_002539_020 |
0.2196982956 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 20 |
Hb_012565_040 |
0.2225133074 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |