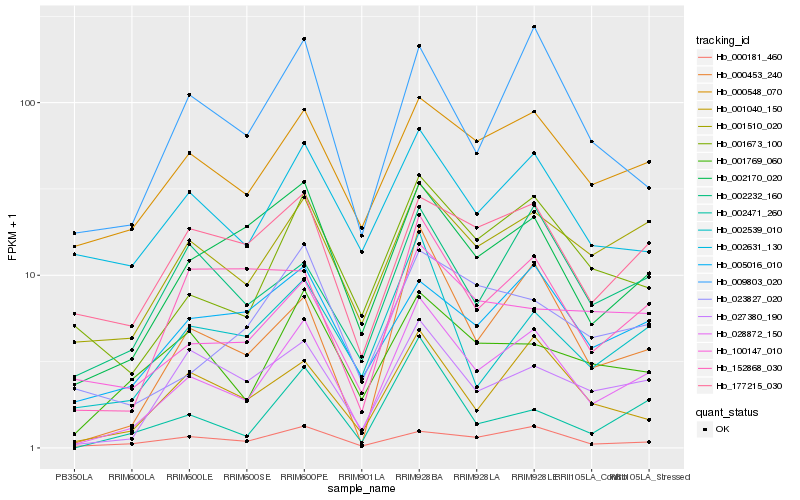
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028872_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |
| 2 |
Hb_000453_240 |
0.1333777142 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 3 |
Hb_001673_100 |
0.1556705019 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 4 |
Hb_002170_020 |
0.1596366738 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 5 |
Hb_001510_020 |
0.1633313026 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 6 |
Hb_027380_190 |
0.1634617112 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001040_150 |
0.1646700773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000548_070 |
0.1706765324 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 9 |
Hb_000181_460 |
0.1724337342 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 10 |
Hb_002539_010 |
0.1772255332 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 11 |
Hb_002471_260 |
0.1773699834 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 12 |
Hb_009803_020 |
0.1818069098 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 13 |
Hb_152868_030 |
0.1833528224 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 14 |
Hb_001769_060 |
0.1842705329 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005016_010 |
0.1860735524 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 16 |
Hb_002631_130 |
0.1866856123 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 17 |
Hb_023827_020 |
0.1886482742 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 18 |
Hb_002232_160 |
0.1889030765 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 19 |
Hb_177215_030 |
0.1897863007 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_100147_010 |
0.1908656761 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |