| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
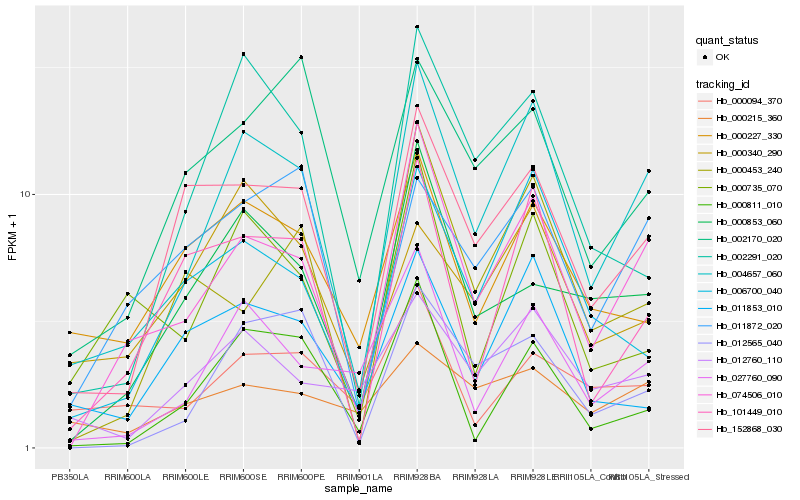
Hb_004657_060 |
0.0 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_074506_010 |
0.1168241445 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 3 |
Hb_152868_030 |
0.1678650893 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 4 |
Hb_006700_040 |
0.1761217982 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 5 |
Hb_027760_090 |
0.180089833 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_012760_110 |
0.1807850066 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002291_020 |
0.1812629155 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_012565_040 |
0.1850736975 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 9 |
Hb_101449_010 |
0.189074316 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 10 |
Hb_011872_020 |
0.194578442 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 11 |
Hb_000735_070 |
0.1955282295 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 12 |
Hb_000853_060 |
0.1956226679 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000227_330 |
0.2000598671 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 14 |
Hb_011853_010 |
0.2005640797 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 15 |
Hb_002170_020 |
0.202101752 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 16 |
Hb_000453_240 |
0.2056814844 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 17 |
Hb_000215_360 |
0.2068658199 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 18 |
Hb_000094_370 |
0.2100537108 |
- |
- |
- |
| 19 |
Hb_000811_010 |
0.2104197258 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 20 |
Hb_000340_290 |
0.2110584359 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |