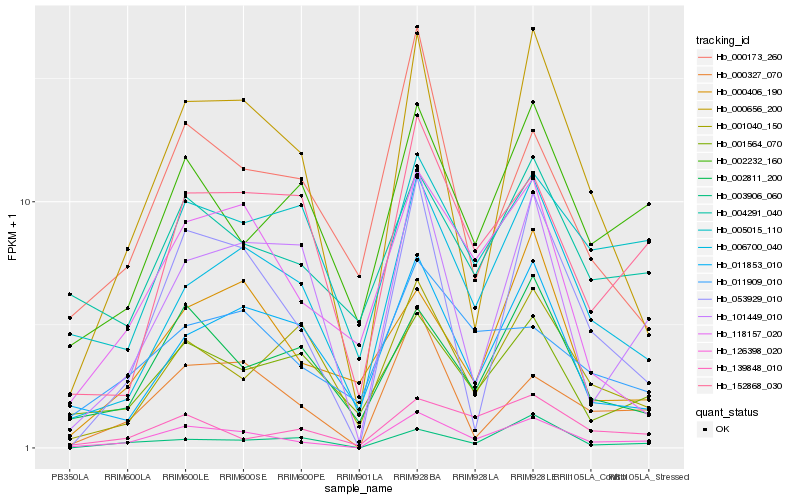
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_126398_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006700_040 |
0.1628897773 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 3 |
Hb_053929_010 |
0.1763968258 |
- |
- |
hypothetical protein POPTR_0010s22320g [Populus trichocarpa] |
| 4 |
Hb_000327_070 |
0.179501825 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000656_200 |
0.1814793945 |
- |
- |
PREDICTED: uncharacterized protein LOC105631373 [Jatropha curcas] |
| 6 |
Hb_152868_030 |
0.1912866778 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 7 |
Hb_002232_160 |
0.1934935106 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 8 |
Hb_002811_200 |
0.1949227285 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 9 |
Hb_118157_020 |
0.1970302276 |
- |
- |
hypothetical protein MTR_3g456110 [Medicago truncatula] |
| 10 |
Hb_101449_010 |
0.19807417 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 11 |
Hb_139848_010 |
0.199642699 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |
| 12 |
Hb_004291_040 |
0.2043686562 |
- |
- |
hexokinase [Manihot esculenta] |
| 13 |
Hb_011853_010 |
0.2045391159 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 14 |
Hb_001564_070 |
0.2058748884 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 15 |
Hb_011909_010 |
0.2076076462 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 16 |
Hb_001040_150 |
0.2135901387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000173_260 |
0.2155639251 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 18 |
Hb_003906_060 |
0.2160702534 |
- |
- |
hypothetical protein VITISV_022975 [Vitis vinifera] |
| 19 |
Hb_005015_110 |
0.2161034495 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 20 |
Hb_000406_190 |
0.2197677229 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |