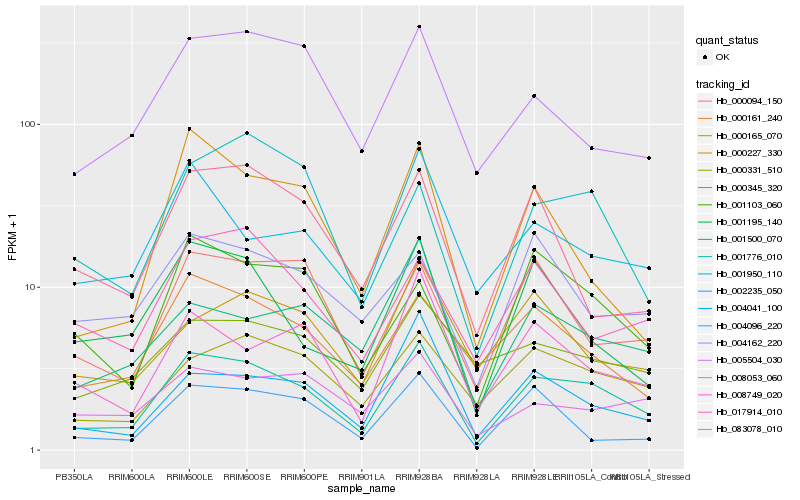
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001776_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 2 |
Hb_000331_510 |
0.1583328301 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 3 |
Hb_000161_240 |
0.1624332305 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_008749_020 |
0.1656362329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004041_100 |
0.1666354589 |
- |
- |
- |
| 6 |
Hb_000094_150 |
0.1682826215 |
- |
- |
- |
| 7 |
Hb_001195_140 |
0.1698320221 |
- |
- |
PREDICTED: U-box domain-containing protein 5-like [Jatropha curcas] |
| 8 |
Hb_001103_060 |
0.17165629 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 9 |
Hb_001500_070 |
0.1733255414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002235_050 |
0.1750628238 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005504_030 |
0.177355885 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 12 |
Hb_000345_320 |
0.1774277792 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_017914_010 |
0.1794782978 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 14 |
Hb_008053_060 |
0.1798500314 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000165_070 |
0.1805220204 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 16 |
Hb_001950_110 |
0.1813400682 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 17 |
Hb_004096_220 |
0.1820710789 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004162_220 |
0.1830149665 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000227_330 |
0.1834259661 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 20 |
Hb_083078_010 |
0.1858949931 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |