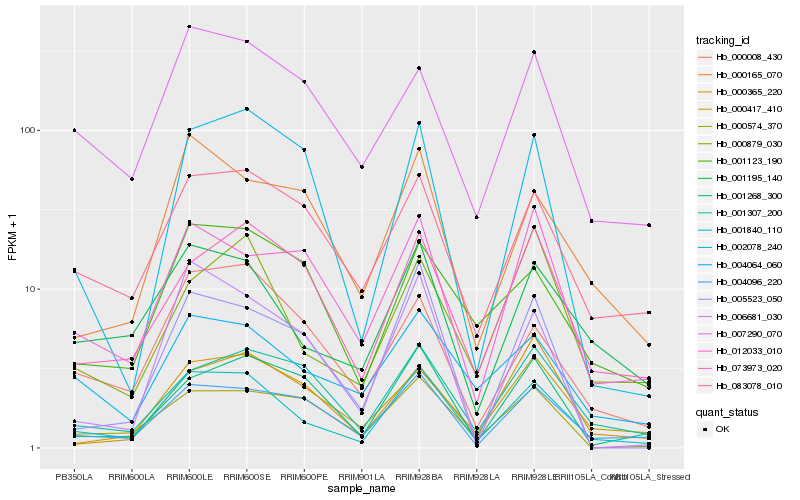
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001840_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 2 |
Hb_001195_140 |
0.1416094826 |
- |
- |
PREDICTED: U-box domain-containing protein 5-like [Jatropha curcas] |
| 3 |
Hb_004096_220 |
0.1504229573 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 4 |
Hb_083078_010 |
0.1611304622 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 5 |
Hb_004064_060 |
0.1628382944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_073973_020 |
0.1677826282 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 7 |
Hb_000008_430 |
0.1683105406 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000879_030 |
0.173793566 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 9 |
Hb_002078_240 |
0.1738759043 |
rubber biosynthesis |
Gene Name: SRPP4 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 10 |
Hb_000165_070 |
0.1748425581 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 11 |
Hb_007290_070 |
0.1752073554 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 12 |
Hb_001307_200 |
0.1759429443 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 13 |
Hb_006681_030 |
0.177305013 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_001268_300 |
0.1773375192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001123_190 |
0.1789128259 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 16 |
Hb_012033_010 |
0.1814787904 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 17 |
Hb_005523_050 |
0.1815987933 |
- |
- |
hypothetical protein JCGZ_09175 [Jatropha curcas] |
| 18 |
Hb_000574_370 |
0.1817058046 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 19 |
Hb_000365_220 |
0.1851822705 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 20 |
Hb_000417_410 |
0.1867553689 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |