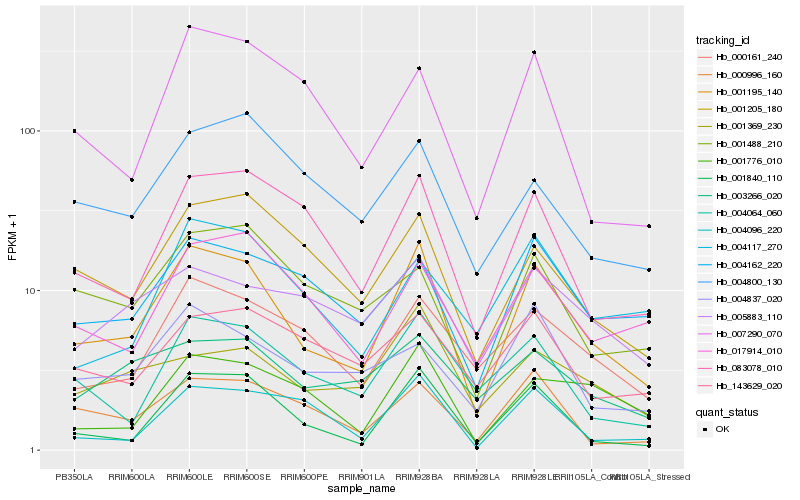
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_140 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 5-like [Jatropha curcas] |
| 2 |
Hb_001840_110 |
0.1416094826 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 3 |
Hb_083078_010 |
0.1581300035 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 4 |
Hb_000161_240 |
0.1627170966 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004064_060 |
0.1646009033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001205_180 |
0.1687610944 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 7 |
Hb_001776_010 |
0.1698320221 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 8 |
Hb_000996_160 |
0.1724452265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004162_220 |
0.1727167554 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_017914_010 |
0.1745127094 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 11 |
Hb_004096_220 |
0.1761425497 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001369_230 |
0.1803275631 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 13 |
Hb_007290_070 |
0.1819119386 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 14 |
Hb_004837_020 |
0.1853895446 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |
| 15 |
Hb_003266_020 |
0.1858680021 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 16 |
Hb_005883_110 |
0.1863360699 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001488_210 |
0.1892072928 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 18 |
Hb_004800_130 |
0.1893804668 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 19 |
Hb_004117_270 |
0.1894680375 |
- |
- |
- |
| 20 |
Hb_143629_020 |
0.1910639608 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |