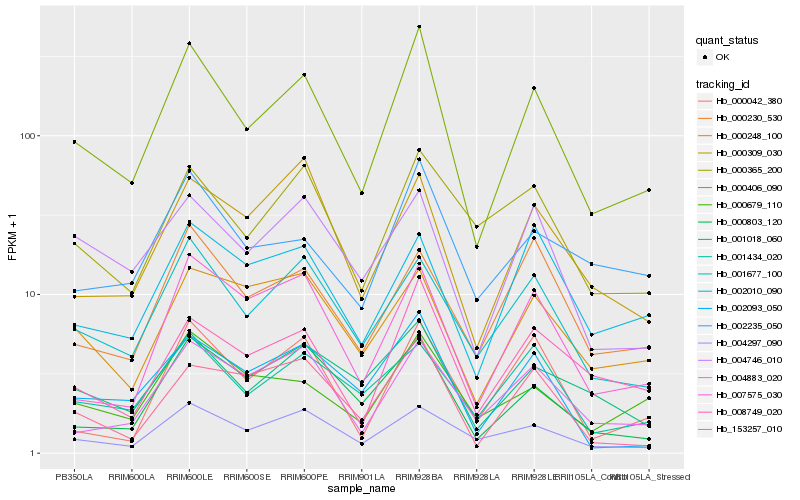
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000406_090 |
0.0 |
- |
- |
unknown [Populus trichocarpa] |
| 2 |
Hb_008749_020 |
0.1302450993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001677_100 |
0.1328807998 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 4 |
Hb_004297_090 |
0.1340257179 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 5 |
Hb_002093_050 |
0.1419632956 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 6 |
Hb_001018_060 |
0.1422291549 |
- |
- |
hypothetical protein B456_001G028000 [Gossypium raimondii] |
| 7 |
Hb_000679_110 |
0.1472983357 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 8 |
Hb_000803_120 |
0.1475244103 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004883_020 |
0.1477564819 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 10 |
Hb_007575_030 |
0.1485082645 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 11 |
Hb_002235_050 |
0.1498444413 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001434_020 |
0.1514517786 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 13 |
Hb_004746_010 |
0.1531063804 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 14 |
Hb_153257_010 |
0.1532333116 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000365_200 |
0.1582079585 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 16 |
Hb_000248_100 |
0.1602004137 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 17 |
Hb_000309_030 |
0.1613315359 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000230_530 |
0.1622024576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002010_090 |
0.1636074465 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000042_380 |
0.1651921354 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |